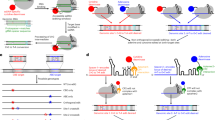
Abstract
Here, we describe an extension of our original transformation-associated recombination (TAR) cloning protocol, enabling selective isolation of DNA segments from microbial genomes. The technique is based on the previously described TAR cloning procedure developed for isolation of a desirable region from mammalian genomes that are enriched in autonomously replicating sequence (ARS)-like sequences, elements that function as the origin of replication in yeast. Such sequences are not common in microbial genomes. In this Protocol Extension, an ARS is inserted into the TAR vector along with a counter-selectable marker, allowing for selection of cloning events against vector circularization. Pre-treatment of microbial DNA with CRISPR–Cas9 to generate double-stranded breaks near the targeted sequences greatly increases the yield of region-positive colonies. In comparison to other available methods, this Protocol Extension allows selective isolation of any region from microbial genomes as well as from environmental DNA samples. The entire procedure can be completed in 10 d.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Kouprina, N. & Larionov, V. TAR cloning: insights into gene function, long-range haplotypes and genome structure and evolution. Nat. Rev. Genet. 7, 805–812 (2006).
Kouprina, N. & Larionov, V. Transformation-associated recombination (TAR) cloning for genomics studies and synthetic biology. Chromosoma 125, 621–632 (2016).
Kouprina, N. & Larionov, V. TAR cloning: perspectives for functional genomics, biomedicine, and biotechnology. Mol. Ther. Methods Clin. Dev. 14, 16–26 (2019).
Kouprina, N. et al. Accelerated evolution of the ASPM gene controlling brain size begins prior to human brain expansion. PLOS Biol. 2, 653–663 (2004).
Kouprina, N. et al. Evolutionary diversification of SPANX-N sperm protein gene structure and expression. PLOS One 2, e359 (2007).
Pavlicek, A. et al. Evolution of the tumor suppressor BRCA1 locus in primates: implications for cancer predisposition. Hum. Mol. Genet. 13, 1–15 (2004).
Lee, N. C. O., Larionov, V. & Kouprina, N. Highly efficient CRISPR/Cas9-mediated TAR cloning of genes and chromosomal loci from complex genomes in yeast. Nucleic Acids Res. 43, e55 (2015).
Theis, J. F. & Newlon, C. S. The ARS309 chromosomal replicator of Saccharomyces cerevisiae depends on an exceptional ARS consensus sequence. Proc. Natl Acad. Sci. USA 94, 10786–10791 (1997).
Stinchcomb, D. T., Thomas, M., Kelly, J., Selker, E. & Davis, R. W. Eukaryotic DNA segments capable of autonomous replication in yeast. Proc. Natl Acad. Sci. USA 77, 4559–4563 (1980).
Choi, S. S. et al. Genome engineering for microbial natural product discovery. Curr. Opin. Microbiol. 45, 53–60 (2018).
Zhang, M. M., Qiao, Y., Ang, E. L. & Zhao, H. Using natural products for drug discovery: the impact of the genomics era. Expert Opin. Drug Discov. 12, 475–487 (2017).
Kouprina, N. & Larionov, V. Selective isolation of genomic loci from complex genomes by transformation-associated recombination cloning in the yeast Saccharomyces cerevisiae. Nat. Protoc. 3, 371–377 (2008).
Noskov, V. N. et al. A general transformation-associated recombination cloning system to selectively isolate any eukaryotic or prokaryotic genomic region. BMC Genomics 4, 16 (2003). Epub 2003 Apr 29.
Zhang, J. J., Yamanaka, K., Tang, X. & Moore, B. S. Direct cloning and heterologous expression of natural product biosynthetic gene clusters by transformation-associated recombination. Methods Enzymol. 621, 87–110 (2019).
Feng, Z., Kim, J. H. & Brady, S. F. Fluostatins produced by the heterologous expression of a TAR reassembled environmental DNA derived type II PKS gene cluster. J. Am. Chem. Soc. 132, 11902–11913 (2010).
Kim, J. H. et al. Cloning large natural product gene clusters from the environment: piecing environmental DNA gene clusters back together with TAR. Biopolymers 93, 833–844 (2010).
Yamanaka, K. et al. Direct cloning and refactoring of a silent lipopeptide biosynthetic gene cluster yields the antibiotic taromycin A. Proc. Natl Acad. Sci. USA 111, 1957–1962 (2014).
Noskov, V. N. et al. Isolation of circular yeast artificial chromosomes for synthetic biology and functional genomics studies. Nat. Protoc. 6, 89–96 (2011).
Gibson, D. G. et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods 6, 343–347 (2009).
Lartigue, C. et al. Creating bacterial strains from genomes that have been cloned and engineered in yeast. Science 325, 1693–1696 (2009).
Benders, G. A. et al. Cloning whole bacterial genomes in yeast. Nucleic Acids Res. 38, 2558–2569 (2010).
Karas, B. J., Tagwerker, C., Yonemoto, I. T., Hutchison, C. A. 3rd & Smith, H. O. Cloning the Acholeplasma laidlawii PG-8A genome in Saccharomyces cerevisiae as a yeast centromeric plasmid. ACS Synth. Biol. 1, 22–28 (2012).
Tagwerker, C. et al. Sequence analysis of a complete 1.66 Mb Prochlorococcus marinus MED4 genome cloned in yeast. Nucleic Acids Res 40, 10375–1038 (2012).
Karas, B. J. et al. Assembly of eukaryotic algal chromosomes in yeast. J. Biol. Eng. 7, 30 (2013).
Mitchell, L. A. et al. Versatile genetic assembly system (VEGAS) to assemble pathways for expression in S. cerevisiae. Nucleic Acids Res. 43, 6620–6630 (2015).
Shang, Y. et al. Construction and rescue of a functional synthetic baculovirus. ACS Synth. Biol. 6, 1393–1402 (2017).
Oldfield, L. M. et al. Genome-wide engineering of an infectious clone of herpes simplex virus type 1 using synthetic genomics assembly methods. Proc. Natl Acad. Sci. USA 114, E8885–E8894 (2017).
Vashee, S. et al. Cloning, assembly, and modification of the primary human cytomegalovirus isolate Toledo by yeast-based transformation-associated recombination. mSphere 2, pii: e00331-17 (2017).
Xie, Y. G. et al. Cloning of a novel, anonymous gene from a megabase-range YAC contig in the neurofibromatosis type 2/meningioma region on human chromosome 22q12. Hum. Mol. Genet. 2, 1361–1368 (1993).
Loots, G. G. Modifying yeast artificial chromosomes to generate Cre/LoxP and FLP/FRT site-specific deletions and inversions. Methods Mol. Biol. 349, 75–84 (2006).
Jiang, W. et al. Cas9-Assisted Targeting of CHromosome segments CATCH enables one-step targeted cloning of large gene clusters. Nat. Commun. 6, 8101 (2015).
Jiang, W. & Zhu, T. F. Targeted isolation and cloning of 100-kb microbial genomic sequences by Cas9-assisted targeting of chromosome segments. Nat. Protoc. 11, 960–975 (2016).
Medema, M. H. & Osbourn, A. Computational genomic identifcation and functional reconstitution of plant natural product biosynthetic pathways. Nat. Prod. Rep. 33, 951–962 (2016).
Noskov, V. N. et al. A novel strategy for analysis of gene homologs and segmental genome duplications. J. Mol. Evol. 56, 702–710 (2003).
Furter-Graves, E. & Hall, B. D. DNA sequence elements required for transcription initiation of the Shizosaccharomyces pombe ADH gene in Saccharomyces cerevisiae. Mol. Gen. Genet. 223, 407–417 (1990).
Miret, J. J., Pessoa-Brandao, L. & Lahue, R. S. Orientation-dependent and sequence-specific expansions of CTG/CAG trinucleotide repeats in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA 95, 12438–12443 (1998).
Noskov, V. et al. Defining the minimal length of sequence homology required for selective gene isolation by TAR cloning. Nucleic Acids Res. 29, E32 (2001).
D’Rose, V., Johny, T. K. & Bhat, S. Comparative analysis of metagenomic DNA extraction methods from gut microbiota of zebrafish (Danio rerio) for downstream next-generation sequencing. J. Appl. Biol. Biotechnol. 7, 1–15 (2019).
Kouprina, N., Noskov, V. N., Koriabine, M., Leem, S. H. & Larionov, V. Exploring transformation-associated recombination cloning for selective isolation of genomic regions. Methods Mol. Biol. 255, 69–89 (2004).
Kouprina, N., Noskov, V. N. & Larionov, V. Selective isolation of large chromosomal regions by transformation-associated recombination cloning for structural and functional analysis of mammalian genomes. Methods Mol. Biol. 349, 85–101 (2006).
Leem, S. H. et al. Optimum conditions for selective isolation of genes from complex genomes by transformation-associated recombination cloning. Nucleic Acids Res. 31, e29 (2003).
Tang, X. et al. Identification of thiotetronic acid antibiotic biosynthetic pathways by target-directed genome mining. ACS Chem. Biol. 10, 2841–2849 (2015).
Bilyk, O., Sekurova, O. N., Zotchev, S. B. & Luzhetskyy, A. Cloning and heterologous expression of the grecocycline biosynthetic gene cluster. PLOS One 11, e0158682 (2016).
Tanveer, A., Yadav, S. & Yadav, D. Comparative assessment of methods for metagenomic DNA isolation from soils of different crop growing fields. 3 Biotech 6, 220 (2016).
Lazarevic, V., Gaïa, N., Girard, M., François, P. & Schrenzel, J. Comparison of DNA extraction methods in analysis of salivary bacterial communities. PLOS One 8, e67699 (2013).
Larionov, V., Kouprina, N., Solomon, G., Barrett, J. C. & Resnick, M. A. Direct isolation of human BRCA2 gene by transformation-associated recombination in yeast. Proc. Natl Acad. Sci. USA 94, 7384–7387 (1997).
Annab, L. et al. Isolation of a functional copy of the human BRCA1 gene by TAR cloning in yeast. Gene 250, 201–208 (2000).
Kouprina, N. et al. Dynamic structure of the SPANX gene cluster mapped to the prostate cancer susceptibility locus HPCX at Xq27. Genome Res. 15, 1477–1486 (2005).
Acknowledgements
This work was supported by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, USA.
Author information
Authors and Affiliations
Contributions
V.L., V.N.N. and N.K. designed the research and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Peer review information Nature Protocols thanks Yinhua Lu and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key references using this protocol
Kouprina, N. and Larionov, V. Mol. Ther. Methods Clin. Dev. 14, 16–26 (2019): https://doi.org/10.1016/j.omtm.2019.05.006
Lee, N. C. O., Larionov, V. and Kouprina, N. Nucleic Acids Res. 43, e55 (2015): https://doi.org/10.1093/nar/gkv112
Kouprina, N. and Larionov, V. Nat. Protoc. 3, 371–377 (2008): https://doi.org/10.1038/nprot.2008.5
Noskov, V. N. et al. BMC Genomics 4, 16 (2003): https://doi.org/10.1186/1471-2164-4-16
This protocol is an extension to: Nat. Protoc. doi:10.1038/nprot.2008.5.
Supplementary information
Rights and permissions
About this article
Cite this article
Kouprina, N., Noskov, V.N. & Larionov, V. Selective isolation of large segments from individual microbial genomes and environmental DNA samples using transformation-associated recombination cloning in yeast. Nat Protoc 15, 734–749 (2020). https://doi.org/10.1038/s41596-019-0280-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0280-1
This article is cited by
-
Cirsiliol induces autophagy and mitochondrial apoptosis through the AKT/FOXO1 axis and influences methotrexate resistance in osteosarcoma
Journal of Translational Medicine (2023)
-
An efficient method for targeted cloning of large DNA fragments from Streptomyces
Applied Microbiology and Biotechnology (2023)
-
Neuroinvasion and anosmia are independent phenomena upon infection with SARS-CoV-2 and its variants
Nature Communications (2023)
-
Cas12a-assisted precise targeted cloning using in vivo Cre-lox recombination
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.