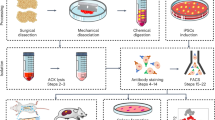
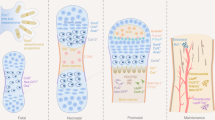
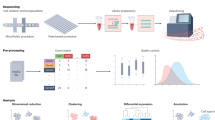
Abstract
Advances in single-cell technologies have transformed the ability to identify the individual cell types present within tissues and organs. The musculoskeletal bionetwork, part of the wider Human Cell Atlas project, aims to create a detailed map of the healthy musculoskeletal system at a single-cell resolution throughout tissue development and across the human lifespan, with complementary generation of data from diseased tissues. Given the prevalence of musculoskeletal disorders, this detailed reference dataset will be critical to understanding normal musculoskeletal function in growth, homeostasis and ageing. The endeavour will also help to identify the cellular basis for disease and lay the foundations for novel therapeutic approaches to treating diseases of the joints, soft tissues and bone. Here, we present a Roadmap delineating the critical steps required to construct the first draft of a human musculoskeletal cell atlas. We describe the key challenges involved in mapping the extracellular matrix-rich, but cell-poor, tissues of the musculoskeletal system, outline early milestones that have been achieved and describe the vision and directions for a comprehensive musculoskeletal cell atlas. By embracing cutting-edge technologies, integrating diverse datasets and fostering international collaborations, this endeavour has the potential to drive transformative changes in musculoskeletal medicine.
Key points
-
Musculoskeletal diseases present a large, and growing, global burden and delivery of effective treatments for these diseases requires improved understanding of the cellular basis of musculoskeletal tissue health and disease.
-
The advent of single-cell sequencing methods allows us to build a ‘human cell atlas’ that identifies and maps every cell of the human body.
-
The application of single-cell technologies in the study of musculoskeletal tissue has historically been difficult owing to the low cellularity and extracellular matrix-rich composition of these tissues.
-
Advances in laboratory and computational methodologies and sharing of knowledge and data among the research community should make it possible to deliver a musculoskeletal cell atlas.
-
Future expansion of the atlas, incorporating a greater number of ancestrally diverse donors, more tissue types and life stages, and diseased samples, will help to accelerate the development of improved diagnostic and therapeutic capabilities.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lee, R. C. et al. Total-body skeletal muscle mass: development and cross-validation of anthropometric prediction models. Am. J. Clin. Nutr. 72, 796–803 (2000).
Murphy, A. C. et al. Structure, function, and control of the human musculoskeletal network. PLoS Biol. 16, e2002811 (2018).
McKee, T. J., Perlman, G., Morris, M. & Komarova, S. V. Extracellular matrix composition of connective tissues: a systematic review and meta-analysis. Sci. Rep. 9, 10542 (2019).
Konnaris, M. A. et al. Computational pathology for musculoskeletal conditions using machine learning: advances, trends, and challenges. Arthritis Res. Ther. 24, 68 (2022).
Freemont, A. J. & Hoyland, J. A. Morphology, mechanisms and pathology of musculoskeletal ageing. J. Pathol. 211, 252–259 (2007).
Sebbag, E. et al. The world-wide burden of musculoskeletal diseases: a systematic analysis of the World Health Organization Burden of Diseases Database. Ann. Rheum. Dis. 78, 844–848 (2019).
Dai, X. & Shen, L. Advances and trends in omics technology development. Front. Med. 9, 911861 (2022).
Regev, A. et al. The human cell atlas. eLife 6, e27041 (2017).
Zeng, H. What is a cell type and how to define it? Cell 185, 2739–2755 (2022).
Stuart, T. & Satija, R. Integrative single-cell analysis. Nat. Rev. Genet. 20, 257–272 (2019).
Moses, L. & Pachter, L. Museum of spatial transcriptomics. Nat. Methods 19, 534–546 (2022).
Vickovic, S. et al. High-definition spatial transcriptomics for in situ tissue profiling. Nat. Methods 16, 987–990 (2019).
Efremova, M., Vento-Tormo, M., Teichmann, S. A. & Vento-Tormo, R. CellPhoneDB: inferring cell–cell communication from combined expression of multi-subunit ligand–receptor complexes. Nat. Protoc. 15, 1484–1506 (2020).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 24, 550–572 (2023).
Cieza, A. et al. Global estimates of the need for rehabilitation based on the Global Burden of Disease study 2019: a systematic analysis for the Global Burden of Disease Study 2019. Lancet 396, 2006–2017 (2020).
World Health Organization. Musculoskeletal Health. Who.int https://www.who.int/news-room/fact-sheets/detail/musculoskeletal-conditions (2022).
Briggs, A. M. et al. Musculoskeletal health conditions represent a global threat to healthy aging: a report for the 2015 World Health Organization World Report on Ageing and Health. Gerontologist 56, S243–S255 (2016).
Staal, J. B., de Bie, R., de Vet, H. C. W., Hildebrandt, J. & Nelemans, P. Injection therapy for subacute and chronic low‐back pain. Cochrane Database Syst. Rev. 2008, CD001824 (2008).
Beard, D. J. et al. Arthroscopic subacromial decompression for subacromial shoulder pain (CSAW): a multicentre, pragmatic, parallel group, placebo-controlled, three-group, randomised surgical trial. Lancet 391, 329–338 (2018).
Jüni, P. et al. Intra-articular corticosteroid for knee osteoarthritis. Cochrane Database Syst. Rev. 2015, CD005328 (2015).
Qiu, X. et al. Single-cell RNA sequencing of human femoral head in vivo. Aging 13, 15595–15619 (2021).
Kendal, A. R. et al. Multi-omic single cell analysis resolves novel stromal cell populations in healthy and diseased human tendon. Sci. Rep. 10, 13939 (2020).
Akbar, M. et al. Single cell and spatial transcriptomics in human tendon disease indicate dysregulated immune homeostasis. Ann. Rheum. Dis. 80, 1494–1497 (2021).
Zhao, H. et al. Single-cell transcriptomics analysis of the pathogenesis of tendon injury. Oxid. Med. Cell Longev. 2022, 7887782 (2022).
Karlsen, A. et al. Distinct myofibre domains of the human myotendinous junction revealed by single-nucleus RNA sequencing. J. Cell Sci. 136, jcs.260913 (2023).
Fu, W., Yang, R. & Li, J. Single-cell and spatial transcriptomics reveal changes in cell heterogeneity during progression of human tendinopathy. BMC Biol. 21, 132 (2023).
Gan, Y. et al. Spatially defined single-cell transcriptional profiling characterizes diverse chondrocyte subtypes and nucleus pulposus progenitors in human intervertebral discs. Bone Res. 9, 37 (2021).
Jiang, W. et al. Single-cell atlas unveils cellular heterogeneity and novel markers in human neonatal and adult intervertebral discs. iScience 25, 104504 (2022).
Cherif, H. et al. Single-cell RNA-seq analysis of cells from degenerating and non-degenerating intervertebral discs from the same individual reveals new biomarkers for intervertebral disc degeneration. Int. J. Mol. Sci. 23, 3993 (2022).
Li, Z. et al. Single-cell RNA sequencing reveals the difference in human normal and degenerative nucleus pulposus tissue profiles and cellular interactions. Front. Cell Dev. Biol. 10, 910626 (2022).
Fu, W. et al. Cellular features of localized microenvironments in human meniscal degeneration: a single-cell transcriptomic study. eLife 11, e79585 (2022).
Swahn, H. et al. Senescent cell population with ZEB1 transcription factor as its main regulator promotes osteoarthritis in cartilage and meniscus. Ann. Rheum. Dis. 82, 403–415 (2023).
De Micheli, A. J., Spector, J. A., Elemento, O. & Cosgrove, B. D. A reference single-cell transcriptomic atlas of human skeletal muscle tissue reveals bifurcated muscle stem cell populations. Skelet. Muscle 10, 19 (2020).
Perez, K. et al. Single nuclei profiling identifies cell specific markers of skeletal muscle aging, frailty, and senescence. Aging 14, 9393–9422 (2022).
Lovrić, A. et al. Single-cell sequencing deconvolutes cellular responses to exercise in human skeletal muscle. Commun. Biol. 5, 1121 (2022).
Wang, X. et al. Comparison of the major cell populations among osteoarthritis, Kashin–Beck disease and healthy chondrocytes by single-cell RNA-seq analysis. Cell Death Dis. 12, 551 (2021).
Orchard, P. et al. Human and rat skeletal muscle single-nuclei multi-omic integrative analyses nominate causal cell types, regulatory elements, and SNPs for complex traits. Genome Res. 31, 2258–2275 (2021).
Stephenson, W. et al. Single-cell RNA-seq of rheumatoid arthritis synovial tissue using low-cost microfluidic instrumentation. Nat. Commun. 9, 791 (2018).
Armaka, M. et al. Single-cell multimodal analysis identifies common regulatory programs in synovial fibroblasts of rheumatoid arthritis patients and modeled TNF-driven arthritis. Genome Med. 14, 78 (2022).
Croft, A. P. et al. Distinct fibroblast subsets drive inflammation and damage in arthritis. Nature 570, 246–251 (2019).
Rubenstein, A. B. et al. Single-cell transcriptional profiles in human skeletal muscle. Sci. Rep. 10, 229 (2020).
Piccolo, S. R. & Frampton, M. B. Tools and techniques for computational reproducibility. GigaScience 5, https://doi.org/10.1186/s13742-016-0135-4 (2016).
Perkel, J. M. The sleight-of-hand trick that can simplify scientific computing. Nature 617, 212–213 (2023).
Osumi-Sutherland, D. et al. Cell type ontologies of the human cell atlas. Nat. Cell Biol. 23, 1129–1135 (2021).
Sender, R., Fuchs, S. & Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 14, e1002533 (2016).
Krasniewski, L. K. et al. Single-cell analysis of skeletal muscle macrophages reveals age-associated functional subpopulations. eLife 11, e77974 (2022).
Schonfeldova, B., Zec, K. & Udalova, I. A. Synovial single-cell heterogeneity, zonation and interactions: a patchwork of effectors in arthritis. Rheumatology 61, 913–925 (2022).
Yin, Z. et al. Atlas of musculoskeletal stem cells with the soft and hard tissue differentiation architecture. Adv. Sci. 7, 2000938 (2020).
Xi, H. et al. A human skeletal muscle atlas identifies the trajectories of stem and progenitor cells across development and from human pluripotent stem cells. Cell Stem Cell 27, 158–176.e110 (2020).
Kelly, N. H., Huynh, N. P. T. & Guilak, F. Single cell RNA-sequencing reveals cellular heterogeneity and trajectories of lineage specification during murine embryonic limb development. Matrix Biol. 89, 1–10 (2020).
Tsai, S. L., Nödl, M. T. & Galloway, J. L. Bringing tendon biology to heel: leveraging mechanisms of tendon development, healing, and regeneration to advance therapeutic strategies. Dev. Dyn. 250, 393–413 (2021).
Kehl, A. S., Corr, M. & Weisman, M. H. Review: enthesitis: new insights into pathogenesis, diagnostic modalities, and treatment. Arthritis Rheumatol. 68, 312–322 (2016).
Millar, N. L. et al. Tendinopathy. Nat. Rev. Dis. Prim. 7, 1 (2021).
Bosisio, F. M. et al. Next-generation pathology using multiplexed immunohistochemistry: mapping tissue architecture at single-cell level. Front. Oncol. 12, 918900 (2022).
Grogan, S. P. et al. Zone-specific gene expression patterns in articular cartilage. Arthritis Rheum. 65, 418–428 (2013).
Hellingman, C. A. et al. Differences in cartilage-forming capacity of expanded human chondrocytes from ear and nose and their gene expression profiles. Cell Transplant. 20, 925–940 (2011).
Petrany, M. J. et al. Single-nucleus RNA-seq identifies transcriptional heterogeneity in multinucleated skeletal myofibers. Nat. Commun. 11, 6374 (2020).
Cutler, A. A., Jackson, J. B., Corbett, A. H. & Pavlath, G. K. Non-equivalence of nuclear import among nuclei in multinucleated skeletal muscle cells. J. Cell Sci. 131, jcs207670 (2018).
Teytelman, L., Stoliartchouk, A., Kindler, L. & Hurwitz, B. L. Protocols.io: virtual communities for protocol development and discussion. PLoS Biol. 14, e1002538 (2016).
Lindsay, S. & Copp, A. J. MRC-Wellcome Trust human developmental biology resource: enabling studies of human developmental gene expression. Trends Genet. 21, 586–590 (2005).
Ramos-Mucci, L., Sarmiento, P., Little, D. & Snelling, S. Research perspectives — pipelines to human tendon transcriptomics. J. Orthop. Res. 40, 993–1005 (2022).
Samuelsen, B. T. et al. Hamstring autograft versus patellar tendon autograft for ACL reconstruction: is there a difference in graft failure rate? A meta-analysis of 47,613 patients. Clin. Orthop. Relat. Res. 475, 2459–2468 (2017).
Craig, R. S., Goodier, H., Singh, J. A., Hopewell, S. & Rees, J. L. Shoulder replacement surgery for osteoarthritis and rotator cuff tear arthropathy. Cochrane Database Syst. Rev. 4, CD012879 (2020).
Morris, E. L. et al. Single-cell transcriptomics of suprachiasmatic nuclei reveal a Prokineticin-driven circadian network. EMBO J. 40, e108614 (2021).
Chang, J. et al. Circadian control of the secretory pathway maintains collagen homeostasis. Nat. Cell Biol. 22, 74–86 (2020).
Xu, Z. et al. High-throughput single nucleus total RNA sequencing of formalin-fixed paraffin-embedded tissues by snRandom-seq. Nat. Commun. 14, 2734 (2023).
Mattei, D. et al. Enzymatic dissociation induces transcriptional and proteotype bias in brain cell populations. Int. J. Mol. Sci. 21, 7944 (2020).
Ding, J. et al. Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746 (2020).
Bergen, V., Soldatov, R. A., Kharchenko, P. V. & Theis, F. J. RNA velocity — current challenges and future perspectives. Mol. Syst. Biol. 17, e10282 (2021).
Zhang, X. et al. Comparative analysis of droplet-based ultra-high-throughput single-cell RNA-seq systems. Mol. Cell 73, 130–142.e135 (2019).
Clark, I. C. et al. Microfluidics-free single-cell genomics with templated emulsification. Nat. Biotechnol. https://doi.org/10.1038/s41587-023-01685-z (2023).
Xie, Y. et al. Comparative analysis of single-cell RNA sequencing methods with and without sample multiplexing. Preprint at bioRxiv https://doi.org/10.1101/2023.06.28.546827 (2023).
Luecken, M. D. & Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
GTEx Consortium. The GTEx consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318-1330, (2020).
Dunham, I. et al. An integrated encyclopedia of DNA elements in the human genome. Nature 489, 57–74 (2012).
Habib, N. et al. Massively parallel single-nucleus RNA-seq with DroNc-seq. Nat. Methods 14, 955–958 (2017).
Westendorf, J. J., Bonewald, L. F., Kiel, D. P. & Burtt, N. P. The musculoskeletal knowledge portal: improving access to multi-omics data. Nat. Rev. Rheumatol. 18, 1–2 (2022).
Consortium*, T. T. S. et al. The tabula sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022).
Rood, J. E. et al. Toward a common coordinate framework for the human body. Cell 179, 1455–1467 (2019).
Björnsson, B. et al. Digital twins to personalize medicine. Genome Med. 12, 4 (2019).
Osanlouy, M. et al. The SPARC DRC: building a resource for the autonomic nervous system community. Front. Physiol. 12, 693735 (2021).
Le Rochais, M., Hemon, P., Pers, J. O. & Uguen, A. Application of high-throughput imaging mass cytometry hyperion in cancer research. Front. Immunol. 13, 859414 (2022).
Khoury, B. M. et al. The use of nano-computed tomography to enhance musculoskeletal research. Connect. Tissue Res. 56, 106–119 (2015).
Peyrin, F., Dong, P., Pacureanu, A. & Langer, M. Micro- and nano-CT for the study of bone ultrastructure. Curr. Osteoporos. Rep. 12, 465–474 (2014).
Vanderploeg, E. J., Wilson, C. G. & Levenston, M. E. Articular chondrocytes derived from distinct tissue zones differentially respond to in vitro oscillatory tensile loading. Osteoarthritis Cartilage 16, 1228–1236 (2008).
Khoshgoftar, M., Torzilli, P. A. & Maher, S. A. Influence of the pericellular and extracellular matrix structural properties on chondrocyte mechanics. J. Orthop. Res. 36, 721–729 (2018).
Juras, V., Mlynarik, V., Szomolanyi, P., Valkovič, L. & Trattnig, S. Magnetic resonance imaging of the musculoskeletal system at 7T: morphological imaging and beyond. Top. Magn. Reson. Imaging 28, 125–135 (2019).
Mabbott, N. A. et al. An expression atlas of human primary cells: inference of gene function from coexpression networks. BMC Genomics 14, 632 (2013).
Barret, T. & Edgar, R. in Gene Mapping, Discovery, and Expression Methods in Molecular Biology (ed. Bina, M.) 175–190 (Humana Press, 2006).
Parkinson, H. et al. ArrayExpress — a public database of microarray experiments and gene expression profiles. Nucleic Acid. Res. 35, D747–D750 (2007).
Dave, M., Rankin, J., Pearce, M. & Foster, H. E. Global prevalence estimates of three chronic musculoskeletal conditions: club foot, juvenile idiopathic arthritis and juvenile systemic lupus erythematosus. Pediatr. Rheumatol. 18, 49 (2020).
Huang, Q., Liu, Y., Du, Y. & Garmire, L. X. Evaluation of cell type annotation R packages on single-cell RNA-seq data. Genomics Proteom. Bioinforma. 19, 267–281 (2021).
Fu, R. et al. clustifyr: an R package for automated single-cell RNA sequencing cluster classification. F1000Res 9, 223 (2020).
Lin, Y. et al. scClassify: sample size estimation and multiscale classification of cells using single and multiple reference. Mol. Syst. Biol. 16, e9389 (2020).
Ding, J. & Regev, A. Deep generative model embedding of single-cell RNA-Seq profiles on hyperspheres and hyperbolic spaces. Nat. Commun. 12, 2554 (2021).
Schaum, N. et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature 562, 367–372 (2018).
Sikkema, L. et al. An integrated cell atlas of the lung in health and disease. Nat. Med. 29, 1563–1577 (2023).
Herpelinck, T. et al. An integrated single-cell atlas of the skeleton from development through adulthood. Preprint at bioRxiv https://doi.org/10.1101/2022.03.14.484345 (2022).
Füllgrabe, A. et al. Guidelines for reporting single-cell RNA-seq experiments. Nat. Biotechnol. 38, 1384–1386 (2020).
Siemionow, K., An, H., Masuda, K., Andersson, G. & Cs-Szabo, G. The effects of age, sex, ethnicity, and spinal level on the rate of intervertebral disc degeneration: a review of 1712 intervertebral discs. Spine 36, 1333–1339 (2011).
Owens, B., Mountcastle, S. & White, D. Racial differences in tendon rupture incidence. Int. J. Sports Med. 28, 617–620 (2007).
Yip, K. & Navarro-Millán, I. Racial, ethnic, and healthcare disparities in rheumatoid arthritis. Curr. Opin. Rheumatol. 33, 117–121 (2021).
Buechler, M. B. et al. Cross-tissue organization of the fibroblast lineage. Nature 593, 575–579 (2021).
The Cancer Genome Atlas Research Network et al. The Cancer Genome Atlas Pan-Cancer Analysis project. Nat. Genet 45, 1113–1120 (2013).
Nakayama, K. H., Hou, L. & Huang, N. F. Role of extracellular matrix signaling cues in modulating cell fate commitment for cardiovascular tissue engineering. Adv. Healthc. Mater. 3, 628–641 (2014).
Rowland, C. R., Little, D. & Guilak, F. Factors influencing the long-term behavior of extracellular matrix-derived scaffolds for musculoskeletal soft tissue repair. J. Long. Term. Eff. Med. Implant. 22, 181–193 (2012).
Nikonova, E., Kao, S. Y. & Spletter, M. L. Contributions of alternative splicing to muscle type development and function. Semin. Cell Dev. Biol. 104, 65–80 (2020).
Yanagi, K., Kaname, T., Chinen, Y. & Naritomi, K. Novel alternative splicing of human faciogenital dysplasia 1 gene. Congenit. Anom. 44, 137–141 (2004).
Peffers, M. J. et al. Transcriptome analysis of ageing in uninjured human Achilles tendon. Arthritis Res. Ther. 17, 33 (2015).
Wang, T. et al. Single-cell RNA sequence presents atlas analysis for chondrocytes in the talus and reveals the potential mechanism in coping with mechanical stress. Front. Cell Dev. Biol. 10, 1047119 (2022).
Long, H. et al. Prevalence trends of site-specific osteoarthritis from 1990 to 2019: findings from the Global Burden of Disease Study 2019. Arthritis Rheumatol. 74, 1172–1183 (2022).
Finckh, A. et al. Global epidemiology of rheumatoid arthritis. Nat. Rev. Rheumatol. 18, 591–602 (2022).
Mease, P. J. et al. Prevalence of rheumatologist-diagnosed psoriatic arthritis in patients with psoriasis in European/North American dermatology clinics. J. Am. Acad. Dermatol. 69, 729–735 (2013).
Scotti, L., Franchi, M., Marchesoni, A. & Corrao, G. Prevalence and incidence of psoriatic arthritis: a systematic review and meta-analysis. Semin. Arthritis Rheum. 48, 28–34 (2018).
Dean, L. E. et al. Global prevalence of ankylosing spondylitis. Rheumatology 53, 650–657 (2013).
GBD 2021 Low Back Pain Collaborators. Global, regional, and national burden of low back pain, 1990–2020, its attributable risk factors, and projections to 2050: a systematic analysis of the Global Burden of Disease Study 2021. Lancet Rheumatol. 5, e316–e329 (2023).
Wu, A.-M. et al. Global, regional, and national burden of bone fractures in 204 countries and territories, 1990–2019: a systematic analysis from the Global Burden of Disease Study 2019. Lancet Healthy Longev. 2, e580–e592 (2021).
Hopkins, C. et al. Critical review on the socio-economic impact of tendinopathy. Asia Pac. J. Sports Med. Arthrosc. Rehabil. Technol. 4, 9–20 (2016).
Riel, H., Lindstrøm, C. F., Rathleff, M. S., Jensen, M. B. & Olesen, J. L. Prevalence and incidence rate of lower-extremity tendinopathies in a Danish general practice: a registry-based study. BMC Musculoskelet. Disord. 20, 239 (2019).
Renström, P. A. Eight clinical conundrums relating to anterior cruciate ligament (ACL) injury in sport: recent evidence and a personal reflection. Br. J. Sports Med. 47, 367–372 (2013).
Ethgen, O., Beaudart, C., Buckinx, F., Bruyère, O. & Reginster, J. Y. The future prevalence of sarcopenia in Europe: a claim for public health action. Calcif. Tissue Int. 100, 229–234 (2017).
Shafiee, G. et al. Prevalence of sarcopenia in the world: a systematic review and meta-analysis of general population studies. J. Diabetes Metab. Disord. 16, 21 (2017).
de Villiers, T. J. & Goldstein, S. R. Bone health 2022: an update. Climacteric 25, 1–3 (2022).
Mirabello, L., Troisi, R. J. & Savage, S. A. International osteosarcoma incidence patterns in children and adolescents, middle ages and elderly persons. Int. J. Cancer 125, 229–234 (2009).
Sadykova, L. R. et al. Epidemiology and risk factors of osteosarcoma. Cancer Invest. 38, 259–269 (2020).
Regev, A. et al. The Human Cell Atlas White Paper. arXiv 1810, 05192 (2018).
Acknowledgements
This publication is part of the Human Cell Atlas (HCA): www.humancellatlas.org/publications. The authors would like to thank the Musculoskeletal Bionetwork of the Human Cell Atlas for their discussions and input into this roadmap, particularly T. Herpelinck, L. Geris, P. Tylzanowski, (J.) S. Lawrence, J. Mimpen, C. Cohen, T. Boakye-Serebour and S. Teichmann. A.P.C. is funded by Medical Research Council Career Development Fellowship (MR/V010182/1). S.S. and M.B. are funded by the Chan Zuckerberg Initiative (CZIF2019–002426 and CZIF2021–240342) and supported by the National Institute for Health Research Oxford Biomedical Research Centre.
Author information
Authors and Affiliations
Contributions
S.S., M.B., F.G., A.P.C. and P.H. wrote the article. All authors researched data for the article, contributed substantially to discussion of content and reviewer and/or edited the manuscript before submission.
Corresponding author
Ethics declarations
Competing interests
A.P.C. is listed as an inventor on several patents filed by Oxford University Innovations concerning single-cell sequencing technologies. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Reviews Rheumatology thanks Nidhi Bhutani, Deepak Rao, Gabriela Loots and Hui Shen for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Cellxgene: https://cellxgene.cziscience.com/
GitHub: https://github.com/humancellatlas; https://github.com/
Human Cell Atlas biological networks: https://www.humancellatlas.org/biological-networks/
Human Cell Atlas Data Coordination Platform: https://data.humancellatlas.org/
Human Cell Atlas Metrics Dashboard: https://www.humancellatlas.org/learn-more/hca-metrics/
Human Cell Atlas project: https://www.humancellatlas.org/
Protocols.io: https://www.protocols.io/welcome
The Musculoskeletal Knowledge Portal: https://msk.hugeamp.org/
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Baldwin, M., Buckley, C.D., Guilak, F. et al. A roadmap for delivering a human musculoskeletal cell atlas. Nat Rev Rheumatol 19, 738–752 (2023). https://doi.org/10.1038/s41584-023-01031-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41584-023-01031-2