Abstract
Benefitting from the advantages of high imaging throughput and low cost, wide-field microscopy has become indispensable in biomedical studies. However, it remains challenging to record biodynamics with a large field of view and high spatiotemporal resolution due to the limited space–bandwidth product. Here we propose random-access wide-field (RA-WiFi) mesoscopy for the imaging of in vivo biodynamics over a 163.84 mm2 area with a spatial resolution of ~2.18 μm. We extend the field of view beyond the nominal value of the objective by enlarging the object distance, which leads to a lower field angle, followed by the correction of optical aberrations. We also implement random-access scanning with structured illumination, which enables optical-sectioning capability and high imaging contrast. The multi-plane imaging capability also makes the technique suitable for curved-surface samples. We demonstrate RA-WiFi mesoscopy in multi-modal imaging, including bright-field, dark-field and multi-colour fluorescence imaging. Specifically, we apply RA-WiFi mesoscopy to calcium imaging of cortex-wide neural network activities in awake mice in vivo, under both physiological and pathological conditions. We also show its unique capability in the three-dimensional random access of irregular regions of interest via the biodynamic imaging of mouse spinal cords in vivo. As a compact, low-cost mesoscope with optical-sectioning capability, RA-WiFi mesoscopy will enable broad applications in the biodynamic study of biological systems.
Similar content being viewed by others
Main
The large-scale recording of biodynamics at high spatiotemporal resolution is highly desirable for the study of biological systems1. For example, to study the information interchange and interactions among various areas of the brain in neuroscience, one should monitor these areas simultaneously at single-neuron resolution. Unfortunately, the inherent space–bandwidth product (SBP) of conventional microscopes presents a barrier for this2. As a result, there is a trade-off between the field of view (FOV) and the spatial resolution. Moreover, the data-recording strategy should match the high data throughput, according to the Nyquist sampling theory.
To break the SBP limitation, multiple mesoscopes with custom-designed objectives have been proposed3,4,5,6,7,8,9,10. For example, a bright-field mesoscope with an FOV diameter (⌀) of 40 mm and 1.5 μm spatial resolution as well as mosaic scanning3 was designed, which gives an SBP of 1,422 million—much higher than those of common commercial objectives (for example, XLPLN25XWMP2 (Olympus), SBP = 12.1 million). However, as a wide-field mesoscope, it suffers from a strong out-of-focus background, resulting in a deteriorated image contrast and a shallow imaging depth. To inhibit background pollution, point-scanning methods, including confocal microscopy and two-photon microscopy, can be adopted. For example, a two-photon mesoscope of ⌀ = 5 mm FOV and 0.66 μm spatial resolution4 was designed, with a high SBP of 114 million, for the calcium imaging of neural network activities in the deep brains of mice in vivo. However, the cost of laser scanning is a low imaging speed. To record a 23.76 mm2 FOV at a spatial resolution of 2.6 μm, it took 1.43 s, failing to catch the calcium signals of active neurons. This also suggests that, for the large-scale, high-resolution imaging of biodynamics, it is not sufficient to adopt only custom-designed objectives with high SBPs if no new recording strategy is included.
Recently, a real-time, ultra-large-scale, high-resolution (RUSH) imaging platform with a 120 mm2 FOV, subcellular resolution and a data throughput of 5.1 gigapixels per second was developed5, which utilized an array of 35 scientific complementary metal–oxide–semiconductor (sCMOS) cameras. In addition, a computational microscope with a 135 cm2 FOV, 18 μm resolution and a throughput of 5 gigapixels per second was developed6, which adopted an array of 54 cameras. However, these designs lack the flexibility to avoid recording blank regions and may transfer useless data. Even worse, the system’s complexity and cost will become an important concern for wide adoption in biomedical studies.
Here we propose the random-access wide-field (RA-WiFi) mesoscope, which achieves a large FOV of 163.84 mm2 at a spatial resolution of ~2.18 μm using mostly off-the-shelf optics and ensures a high temporal resolution via the random-access strategy. We extend the FOV of the commercial objective to beyond its nominal value by enlarging the object distance, which subsequently leads to a lower field angle (LFA), and correct optical aberrations using adaptive optics (AO) accompanied with an electronically tunable lens (ETL) for compensating Petzval field curvature. We divide the whole FOV into multiple sub-FOVs for high-resolution imaging. We achieve optical sectioning via structured illumination and computation reconstruction, and demonstrate multiple-plane imaging to match the curved planes of biosamples by adjusting the ETL. In particular, we demonstrate the superior performance of RA-WiFi fluorescence mesoscopy in the cortex-wide imaging of neural network activities in awake mice, under air-puff stimuli and drug-induced epilepsy, respectively. We verify the flexibility of RA-WiFi in allocating limited imaging resources to targeted regions of interest (ROIs), such as the rectangular window of mouse spinal cords, thus avoiding the acquisition of blank regions and the transfer of redundant data. Moreover, we propose the scheme of ‘one shot, multiple sub-FOVs’ to improve the temporal resolution. By reducing the data-readout time, we demonstrate neural imaging at 18.5 Hz across two sub-FOVs at ~25% faster than the camera’s frame-rate limitation.
Results
The principle and system design of RA-WiFi mesoscopy
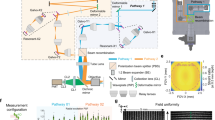
We used a commercial infinity-corrected objective (XLFLUOR4X/340) of SBP = 50 million to build the RA-WiFi mesoscope (Fig. 1a and Supplementary Fig. 1). Considering the system cost, we chose off-the-shelf components, apart from lenses L5 and L6 that were custom-designed for a larger aperture and less aberration. To bridge the huge gap between the whole FOV (supported by the objective) and the small area (supported by the sCMOS camera) in high-resolution imaging, we performed random-access scanning using an xy-galvanometric mirror pair (two-axis galvo). In fluorescence imaging mode, the two-axis galvo steers the excitation light to illuminate different sub-FOVs and performs a de-scan for emission fluorescence signals (Fig. 1b; see also Supplementary Fig. 2). In both bright-field and dark-field imaging modes, we illuminate the whole FOV simultaneously but collect sub-FOV images sequentially by driving the two-axis galvo. The strategy of random-access scanning makes it flexible in imaging samples of various shapes and sizes, with improved temporal resolution in practical imaging.
a, Optical schematic of the RA-WiFi mesoscope. Light emitted from light-emitting-diodes LED1 and LED2 first pass through a dichroic mirror (DM1) and an excitation filter (FEX), and are then delivered to a digital micromirror device (DMD) via a total internal reflection (TIR) prism. The modulated light is then projected on to the sample plane through three sets of relay lenses, which includes the pairs of L1 (focal length (f) = 150 mm) and L2 (f = 150 mm), and L3 (f = 180 mm) and L4 (f = 102 mm), as well as custom-designed lenses L5, L6 and the objective. The second dichroic mirror DM2, the ETL and the two-axis galvo are positioned between L3 and L4. Fluorescence signals are collected by the same objective and first relayed to the surface of a deformable mirror by a relay lens set (consisting of two scan lens (SL1 and SL2) sets (f = 70 mm)) before being imaged on to the camera through an emission filter (FEM) and a tube lens (TL) (f = 180 mm). b, The principle of random-access imaging. By steering the scan mirror, light emitted from different field positions can be deflected to a de-scanned path and captured by the camera. Sub-FOVs accessed via sequential random-access scanning can be assembled into a mosaic image of the larger scene. c, The LFA configuration. Left: diffraction-limited model, with the nominal FOV of the objective. Middle: LFA configuration, with an extended FOV and accompanying aberrations, such as field curvature. Right: LFA configuration with an extended flat FOV, where the field curvature is corrected via fast axial defocusing with the ETL. d, Schematics of (i) the optical-sectioning capability, (ii) the flexible accessing of ROIs in three-dimensions, (iii) the direct imaging of a tilted sample (of angle θ), (iv) the multi-plane imaging of a curved surface and (v–vii) multiple imaging modes that include dual-colour fluorescence (v), bright-field (vi) and dark-field (vii) imaging modes. e, The typical parameters of imaging speed (frame rate) and FOV size that are demonstrated in this paper. Inset: FWHM values of lateral resolution across 60 different sub-FOVs. Each sub-FOV is 1.6 × 1.6 mm.
To extend the FOV beyond the nominal value of the chosen objective (⌀ = 5.5 mm), we utilized the LFA configuration (Supplementary Fig. 3) and achieve an FOV of 163.84 mm2 (⌀ = 12.8 mm), which is 5.4 times larger. Considering Nyquist sampling with the adopted camera, we set each sub-FOV to be 1.6 × 1.6 mm, and the central positions of the sub-FOVs can be flexibly adjusted for various applications. To correct optical aberrations in such an LFA configuration, including field curvature (Fig. 1c middle), we adopted the ETL for fast axial defocusing (Fig. 1c right) and the deformable mirror for sensorless AO correction (Methods). For optimal imaging in each sub-FOV, we first adjusted the ETL to correct the field curvature and then performed wavefront corrections to compensate for residual system aberrations. In fluorescence imaging mode, we also performed structured illumination using a digital micromirror device (DMD) and computational reconstruction (Methods) for optical sectioning (Supplementary Fig. 4) in both in vitro and in vivo applications. In Supplementary Fig. 5, we show that the FOV of our RA-WiFi mesoscope is large enough to cover two brain slices from an adult mouse in a single frame, and the imaging contrast of neural structures is improved 2.3-fold, benefitting from structured illumination and AO correction.
The comprehensive capabilities of the extended FOV, fast axial adjustment, fine wavefront compensation and optical sectioning make the RA-WiFi mesoscope suitable for various biomedical studies. For example, the optical-sectioning capability (Fig. 1d(i)) suppresses the out-of-focus background and thus ensures high-contrast imaging. Besides, the coordination of random-access scanning and fast axial adjustment enables the flexible access of ROIs in three-dimensions (Fig. 1d(ii)) and the multi-plane imaging of non-planar surfaces (Fig. 1d(iii),(iv)). As an example, the matching between the plane of focus and the sample can be easily achieved by adjusting the ETL to fit the tilted plane, as calculated from the measured positions of three non-collinear sub-FOVs (Supplementary Fig. 6). Carrying out complex processes to align tilted samples, as with conventional mesoscopes, is not necessary. As another example, we performed the brain-wide functional imaging of a mouse pup with an intact curved skull (Supplementary Fig. 7). Furthermore, taking advantage of the extended FOV and multi-plane imaging, we carried out the simultaneous imaging of two mouse pups with intact curved skulls (Supplementary Fig. 8).
The RA-WiFi mesoscope supports multiple imaging modes (Fig. 1d(v)–(vii)). As examples of dual-colour fluorescence imaging, we investigated the neurovascular coupling in Thy1-jRGECO1 mice by recording vascular dilations and calcium signals in vivo simultaneously, without and with air-puff stimuli, shown in Supplementary Figs. 9 and 10, respectively. As examples of multi-modal imaging, we captured the bright-field (62.5 Hz for each sub-FOV) and dark-field images of integrated circuits (Supplementary Fig. 11) as well as carrying out the fluorescence and dark-field imaging of cultured HeLa cells that express green fluorescent protein (Supplementary Fig. 12).
The system performance of the RA-WiFi mesoscope in representative applications is shown in Fig. 1e. Based on practical requirements, the RA-WiFi mesoscope provides flexible choices of the imaging speed (frame rate) and ROI size. It should be noted that the RA-WiFi mesoscope is also suited to the imaging of long-span but isolated ROIs at high spatiotemporal resolution. In the inset of Fig. 1e, we show the measured spatial resolution, with the full-width at half-maximum (FWHM) varying from 2.18 to 3.47 μm across 60 sub-FOVs (see also Supplementary Fig. 13) in which the FWHM values are smaller than 3.1 μm for ~82% of the sub-FOVs.
Cortex-wide functional imaging of mouse brains in vivo
The dynamical imaging of large neural ensembles at single-neuron resolution in vivo is crucial for interrogating neural circuits. However, in current reports, only either small FOV imaging with subcellular resolution or cortex-wide imaging with low resolution is achievable11,12. We therefore validated the capability of our RA-WiFi mesoscope in the large-scale recording of neural network activities at high spatiotemporal resolution under physiological and pathological conditions in vivo.
First we performed the in vivo calcium imaging of spontaneous neural activities in GCaMP6f-expressing Ai148D/Rasgrf mice. Benefitting from the flexible random-access capability, we can set the ROIs to match the optical-accessible cranial window and record the activities of ~3,500 neurons over a 6.4 × 6.4 mm FOV at ~1.67 Hz (Fig. 2a,b and Supplementary Video 1). To explore the functional connectivity among individual neurons on a cortex-wide scale, we built a functional network based on correlation coefficients (CCs) between neurons9 (Fig. 2c), where neuron pairs with CCs of >0.4 are regarded as nodes. It shows that linked neuron connections vary from long distances (>4 mm, green lines) to short distances (<1 mm, magenta lines), and representative calcium traces of the correlated neuron pairs are shown in Fig. 2d. On the distance-dependence, we find that the physical distance has little impact on the CCs in the case of spontaneous activity (Fig. 2e), and identify long-distance functional connections with strong correlations (Fig. 2f), which are difficult to be observed using conventional microscopes with small FOVs. We also compared the results obtained via conventional wide-field (c-WiFi) imaging that has no optical-sectioning capability (Supplementary Fig. 14), which shows distance-dependent correlations. This discrepancy may be attributed to the interference from the out-of-focus background, indicating the advantages of the optical-sectioning capability in the RA-WiFi mesoscope.
a–f, Cortex-wide functional imaging of spontaneous neural activity. a, Whole FOV image of a mouse brain cortex (6.4 × 6.4 mm). The dotted orange line outlines the selected ROIs in dynamical imaging carried out at a frame rate of 1.67 Hz. Around 3,500 active neurons are identified. The white dashed lines indicate boundaries of cortical areas. b, Fluorescence signals of calcium dynamics (∆F/F = (F−F0)/F0, the relative increase in fluorescence intensity to baseline F0), corresponding to the neuron segmentations shown as red dots in a. c, Functional network based on the CCs between neurons. Green and magenta lines denote linked neuron connections with distances of >4 mm and <1 mm, respectively. d, Representative examples of calcium signals for long-distance (left) and short-distance (right) correlated neuron pairs. e, Dependence of the CC on the physical distance between neurons. Blue dots indicate neuron-to-neuron pairs and the line shows the linear regression. f, The distribution of CCs with the physical distance between neurons. g–k, Cortex-wide functional imaging of stimuli-induced neural activity. g, Whole FOV image of another mouse brain cortex (6.4 × 6.4 mm). Around 2,300 active neurons are identified. Three ROIs, as outlined by the coloured boxes, are selected for random-access imaging at a 5.12 Hz frame rate. The inset shows the mouse sample with an air-puff (AP) stimulus. h, Representative examples of neural activities from ROIs i, ii and iii in g that are highly correlated with the AP stimuli. i, Representative examples of calcium signals obtained without and with optical sectioning, respectively, from c-WiFi images (blue line, multiplied fourfold for better visualization) and RA-WiFi images (orange line). j, Calcium signals corresponding to neurons in the three ROIs. The orange arrowheads indicate the occurrence of the AP stimuli at 40 s intervals. k, The neuron-to-neuron CCs after hierarchical clustering and sorting. GCaMP6f-expressing Ai148D/Rasgrf mice were used in all experiments.
Moreover, to show the flexibility in monitoring the targeted brain areas at higher frame rates, we performed the random-access imaging of two non-adjacent ROIs at 10 Hz (Supplementary Fig. 15) and three non-adjacent ROIs at 6.7 Hz (Supplementary Fig. 16), with ROIs being located in both brain hemispheres. We also built a functional network of neurons from brain regions spanning 3.5 mm (Supplementary Fig. 15h) and identified pairs of correlated neurons located across the hemispheres. Some hub-like neurons exhibited more links to others over both short and long distances. Besides, the optical-sectioning capability enabled the dual-plane, dual-sub-FOV imaging of four ROIs at 5 Hz (Supplementary Fig. 17). There is no cross-talk for ~90% of neurons from the same lateral sub-FOV but different axial positions, benefitting from the background-inhibition capability of optical sectioning.
We can also interrogate neural activity patterns in awake mice under sensory stimuli. The cortex-wide calcium imaging results with air-puff stimuli are shown in Supplementary Fig. 18 (Supplementary Video 2). At a higher frame rate, we selectively accessed three ROIs at 5.12 Hz (Fig. 2g and Supplementary Video 3). Representative examples of the stimuli-related neural activities are shown in Fig. 2h. Compared with c-WiFi imaging, the contrast of the calcium signals was improved fourfold in RA-WiFi imaging (Fig. 2i and Supplementary Fig. 19), benefitting from suppression of the out-of-focus background. Moreover, some fluorescence peaks (indicated by the dark grey arrow in Fig. 2i), which may be artefacts arising from background fluorescence, show up only in c-WiFi imaging. We arranged the calcium traces sorted in descending order according to the CC (Fig. 2j), and identified stimuli-responsive neurons (neuron contours in Supplementary Fig. 20d–f) and stimuli-related clusters (Fig. 2k).
Epilepsy is a severe neurological disorder that is characterized by recurrent seizures13. Cortex-wide functional imaging provides a new opportunity to visualize the generation and spread of seizures, facilitating the study of epilepsy mechanisms14,15,16,17. We used the RA-WiFi mesoscope to map a seizure spreading throughout the cortex at subcellular resolution in vivo. After inducing epilepsy via the intraperitoneal injection of kainic acid (KA)18, we performed calcium imaging of cortical neurons in both Ai148D/Rasgrf and Ai162/PV mice. Figure 3a,f shows the representative spatiotemporal maps of successive neuron recruitment (Supplementary Videos 4 and 5), where the neuron contours are colour-coded by onset time points in selected seizure events (noted by the arrows in Fig. 3b,g). We can observe weak signals from individual neurons (Supplementary Fig. 21), in addition to the high intensity of global seizure activity, suggesting a large dynamic range of signal acquisition.
a–e, Cortex-wide functional imaging of neural activity during epilepsy in Ai148D/Rasgrf mice. a, Spatiotemporal map of the successive recruitment of Rasgrf-expressing neurons within a 4.8 × 6.4 mm FOV (recorded at 1.67 Hz). Neuron segmentations are colour-coded according to onset time in the selected seizure event, as noted by the arrow in b. b, Calcium signals corresponding to the neuron segmentations in a. c, Neurovascular coupling in seizures. Vascular dilations are measured along the yellow lines shown in a, and the calcium signals are the averages of the fluorescence signals inside the yellow dashed boxes in a. d, Colour-coded seizure-spreading event in a 4.8 × 3.2 mm area (as denoted by the orange dashed box in a), recorded at 3.3 Hz. e, Intensity changes of pixels versus time along line 1 in d. f–j, Cortex-wide functional imaging of neural activity during epilepsy in Ai162/PV mice. f, Spatiotemporal map of successive recruitment of parvalbumin-expressing (PV-expressing) neurons within a 4.8 × 4.8 mm FOV (at 2.8 Hz). Neuron segmentation results are colour-coded according to the onset time in selected seizure events, as denoted by the arrows in g. g, Calcium signals corresponding to the neuron segmentations in f. The upper and lower panels correspond to neural activities in the left and right brain hemispheres, respectively. h, Neurovascular coupling in seizures. Vascular dilations are measured along the yellow lines shown in f, and the calcium signals are the averages of the fluorescence signals inside the yellow dashed boxes in f. i, Colour-coded seizure-spreading events in the 4.8 × 4.8 mm imaging area. j, Intensity changes of pixels versus along line 1 in i.
We also investigated neurovascular coupling during an epileptic seizure. The correlation analysis of blood vessel diameter and local neural activity (Fig. 3c,h) suggests that some blood vessels are prone to constrict significantly when local neurons are recruited to a hyperactive state (Supplementary Fig. 22). The diameter values decrease to minimal values at, respectively, 28.2 ± 8.39 s and 56.48 ± 20.10 s after peak activities of local neurons in the Ai148D/Rasgrf mice (from the averaged value of nine blood vessels in Supplementary Fig. 22) and Ai162/PV mice (from the averaged value of four blood vessels in Fig. 3h).
Furthermore, we analysed the respective seizure propagation patterns in both mouse types for Rasgrf-expressing neurons and PV-expressing interneurons. The epilepsy foci in both brain hemispheres can be clearly identified in both the Ai148D/Rasgrf mice (Fig. 3d, Supplementary Figs. 23 and 24 and Supplementary Video 6) and Ai162/PV mice (Fig. 3i, Supplementary Figs. 25 and 26 and Supplementary Video 7). For the Rasgrf-expressing neurons, seizure recruitments emerge and expand in all directions (Fig. 3d), and pixelwise intensities along orthogonal lines from the epileptic foci rise successively but drop simultaneously once the ictal event ends globally (Fig. 3e and Supplementary Fig. 24c–f). The median lasting time of the hyperactive state varied from 14.7 to 18.6 s (Supplementary Fig. 24b). By contrast, for the PV-expressing interneurons, the seizure spreads mainly towards the middle line (Fig. 3i), and the pixelwise intensities along the plotted lines from the epileptic foci rise and fall in a successive manner (Fig. 3j and Supplementary Fig. 25c–f). The median lasting time of the hyperactive state varies from 41.76 to 54.72 s (Supplementary Fig. 25b), much longer than that of the Rasgrf-expressing neurons.
Flexible imaging of biodynamics in mouse spinal cords in vivo
The spinal cord, as a part of the central nervous system, plays a critical role in the transmission and processing of information. However, limited by available FOVs19, current studies fail to record biodynamics over multiple dorsal spinal segments simultaneously. Even for the latest mesoscopes5,6,20, when considering the rectangular window of a spinal cord (Fig. 4a) there will be a considerable waste in transmitting and saving the data. Moreover, the non-negligible surface curvature of the spinal cord makes it more challenging for dynamical imaging when using conventional microscopes.
a, Photograph of the exposed mouse spinal cord, with the width (W) and length (L) dimensions of the FOV. R, rostral side; C, caudal side. b, The strategy of multi-plane imaging to match the curved surface. Here, four planes at different focal depths are set, which correspond to four sub-FOVs along the spinal cord. c, Single-plane (top, as in conventional microscopes) and multi-plane (bottom) imaging of the mouse spinal cord in vivo, where the blood plasma is stained with FITC–dextran (fluorescein isothiocyanate–dextran). Each image is normalized to the mean intensity (mean(I)) of the corresponding whole FOV. The lower left insets show the Fourier transform (FT) spectra of the whole FOV images (on a log scale), where the dashed white circle denotes the 0.02% strength of the zero frequency. The focal depths of the four planes are labelled alongside the green spots in the bottom image. Scale bar, 500 μm. d, Expanded views of the regions highlighted by the dashed boxes in c. Scale bar, 50 μm. e, High-speed imaging of neutrophil trafficking along blood vessels in the mouse spinal cord in vivo. Whole FOV, 1.7 × 6.5 mm; frame rate, 3.3 Hz. Examples of neutrophil traces are denoted on the temporally averaged grayscale background. Neutrophils move along the traces in the direction marked by the dot, and the moving velocities are colour-coded. The inset shows the geometry of the dorsal venous system. Scale bar, 500 μm. f, Two representative traces of neutrophil trafficking, with red arrows denoting the moving directions. Scale bar, 500 μm. g, Expanded views of the neutrophils at different timestamps in traces 2 and 7, arranged in respective rows. The arrows denote moving directions. The image intensity is normalized to the minimum–maximum range in each timestamp. Scale bar, 50 μm.
The great flexibility of the RA-WiFi mesoscope to allocate sub-FOVs according to sample profiles offers unique advantages in the dynamical imaging of the mouse spinal cord in vivo. To image over a long spinal cord range, we divided the rectangular FOV into four sub-FOVs, where the length of each was adjusted to 1.7 mm to fully cover the width of the laminectomy window (Methods), and carried out multi-plane imaging to match the surface curvature (Fig. 4b). For comparison, we show the FITC-stained vascular images that were acquired both with and without multi-plane imaging of the spinal cord (Fig. 4c). As the surface height varies up to 325 μm, serious blurring exists in the defocused regions for single-plane imaging. In multi-plane imaging, however, both the Fourier analysis and the images of the vessel structure (Fig. 4d) indicate that high-frequency features are maintained. Such a flexible strategy of matching the sample’s specific shape, size and orientation avoids acquiring useless data outside of the targeted ROIs and ensures high-speed recording. As an example, we performed the in vivo imaging of dye perfusion in the blood vessels of the mouse spinal cord at 9.6 Hz over a 6.5 × 1.7 mm FOV (Supplementary Fig. 27 and Supplementary Video 8).
To demonstrate flexible imaging at high spatiotemporal resolutions, we recorded neutrophil trafficking along the superficial blood vessels in the mouse spinal cord in vivo (Fig. 4e). To record neutrophil recruitment induced by tissue damage during laminectomy, we labelled the neutrophils in wild-type C57BL/6 (B6) mice with the Ly6 antibody via retro-orbital injection. We observed various trafficking paths (Fig. 4e, Supplementary Fig. 28 and Supplementary Video 9), such as: (1) travelling along the dorsal spinal vein (dSV) to the rostral or caudal side (traces 1 and 2, respectively); (2) travelling through the dorsal ascending venules (dAVs), followed by moving into the dSV and flowing to the rostral or caudal side (traces 3, 4 and 5); (3) moving from one dAV and travelling a distance of up to hundreds of micrometres in the dSV to another dAV (traces 6 and 7); and (4) moving from the upstream branching venules (UBVs) to the connected dAV (trace 8). In Fig. 4f,g, we show the detailed trafficking paths and timestamps of traces 2 and 7, respectively. The neutrophil in trace 2 migrates >4.9 mm along the dSV, and the neutrophil in trace 7 travels from a dAV to another dAV ~0.8 mm away. Although the average velocity of the trafficking neutrophils is 180 μm s−1, the peak velocity can be >900 μm s−1, which can still be recorded using our RA-WiFi mesoscope.
We also demonstrated the calcium imaging of spontaneous neural activities in a mouse spinal cord in vivo, in which the signals from both single-neuron firing and regional spreading are recorded (Supplementary Fig. 29 and Supplementary Video 10).
Scheme of one-shot, multiple sub-FOVs to speed up imaging
If the exposure time (texpo) can be short enough while ensuring the signal quality, the current speed bottleneck of the RA-WiFi mesoscope is the readout time of our sCMOS camera (15 ms, leading to a maximal frame rate of 67 Hz per sub-FOV). To save time in data transfer and thus speed up the RA-WiFi mesoscope, we propose the scheme of one-shot, multiple sub-FOVs, which takes advantage of the spatiotemporal sparsity of most biological dynamics. In such a scheme, the camera integrates continuous exposures of multiple sub-FOVs but transfers the data only once (Supplementary Fig. 30). With previous information of the spatial distribution, signals can be computationally de-mixed for each sub-FOV.
We demonstrate applications of the ‘one shot, two sub-FOVs’ (1S2F) scheme as examples. We first adopted the 1S2F scheme in the functional imaging of neural activity in Ai148D/Rasgrf mice (Fig. 5a). Using the conventional scheme of ‘one shot, one sub-FOV’ (1S1F) (texpo = 19 ms, 14.7 Hz), we can acquire previous knowledge of the spatial distribution of neurons in two ROIs (Fig. 5b). Using the 1S2F scheme, we can perform functional imaging of the two ROIs (texpo = 19 ms, 18.5 Hz) and record their images in single frames, saving the 15 ms readout time in each period and achieving ~25% faster acquisition. On the basis of the previous information for the neuron contours, we can de-mix the calcium signals (Fig. 5c–e) and calculate the hierarchically clustered neuron-to-neuron CCs (Fig. 5f).
a, Cortex-wide imaging of a Ai148D/Rasgrf mouse brain with a 6.4 × 6.4 mm FOV. The two selected ROIs for 1S2F imaging are denoted by the boxes. b, Superimposed image of neuron segmentations, based on the subsequent 1S1F imaging of ROIs i and ii. c, Neuron segmentations based on 1S2F imaging. d, Calcium signals extracted via 1S2F imaging. The signals in ROIs i and ii can be distinguished. e, Representative neural activities in ROIs i and ii. f, The neuron-to-neuron CCs after hierarchical clustering and sorting. g, Cortex-wide imaging of a B6 mouse brain with FITC-labelled blood plasma in 4.8 × 4.8 mm FOV. The two selected ROIs are denoted by the boxes. h, Colour-coded superimposed morphologies of blood vessels obtained via the subsequent 1S1F imaging of ROIs i and ii. i, Morphology of the blood vessels obtained via 1S2F imaging. j, Relative dilation of blood vessels 1 and 2 along the coloured lines in i. A moving time average filter with a window of ~1 s is applied (solid lines).
We also used the 1S2F scheme for the structural imaging of vascular dynamics in vivo. We used B6 mice with FITC-stained blood plasma21. Similarly, using the 1S1F scheme (texpo = 19 ms, 14.7 Hz) we acquired previous information on the blood vessel distributions in the two ROIs (Fig. 5g,h). Using the 1S2F scheme (texpo = 19 ms, 18.5 Hz), we then recorded images of the two ROIs in a single frame (Fig. 5i). The blood vessels from different ROIs can be identified and measured for variations in their diameter (Fig. 5j).
Discussion
We propose RA-WiFi mesoscopy for the centimetre-scale recording of biodynamics at high spatiotemporal resolution in vivo, which has the capability of optical sectioning to inhibit the out-of-focus background. We validated the superior performance of the RA-WiFi mesoscope through various physiological and pathological experiments in vivo. Based on practical requirements, the RA-WiFi mesoscope offers flexible choices for the imaging speed and imaging regions. For example, we can either record the cortex-wide neural activity for a 6.4 × 6.4 mm FOV at 1.67 Hz or record neural activities in two targeted ROIs spanning 3.5 mm at 10 Hz. Through the demonstration of cortex-wide, subcellular functional imaging during epilepsy, we confirmed that the RA-WiFi mesoscope is capable of monitoring the spatiotemporal seizure patterns of various neuron types. Specifically, we also demonstrated the flexible imaging of biodynamics in mouse spinal cords in vivo, benefitting from the unique capability of the three-dimensional random access of irregular ROIs.
In the demonstrated experiments, the ETL can achieve axial adjustment within the camera’s readout time, thus enabling a fast defocusing capability for flexible control of the axial position of each sub-FOV for the multi-plane imaging of non-planar surfaces. For the successive scanning of sub-FOVs with a larger difference in their focal depths, the longer ETL switching time poses a further speed limitation, although this can be alleviated via hardware optimization22,23.
The current imaging speed is limited by the total time required for light exposure and data readout. With enhanced fluorescence excitation, emission and collection, or using a denoise algorithm, a shorter exposure time is feasible. To reduce the readout time, we proposed the scheme of one-shot, multiple sub-FOVs, and we demonstrated its feasibility through recording neural activity and vascular dilation. With advanced compressive imaging methods24, the camera readout time can be reduced further. Alternatively, deep video interpolation25,26 or event-driven random-access imaging in a large FOV27 can help to overcome the speed limitation.
Even though the sensorless AO is effective in correcting wavefront aberrations, it is generally time-consuming (around tens of minutes) to optimize for suitable compensation. To speed up this process, as an example, we introduced unpaired self-supervised learning to achieve efficient aberration restoration in an optical prior-driven and data-centric manner28,29,30,31. We used a generative adversarial network (GAN) that incorporates patch-based contrastive learning to transform the aberration-distorted images of the cortex-wide mouse brain vascular into aberration-free images, while retaining their structure and content, by utilizing a single image itself (Methods; Supplementary Fig. 31). Based on the less distorted images at central sub-FOVs, our deep-learning approach can restore images of severe aberrations at peripheral sub-FOVs effectively. Overall, the high SBP and flexibility of RA-WiFi mesoscopy make it easier to organically combine state-of-the-art deep-learning methods and optical systems, which may compensate for the imaging trade-offs caused by physical and hardware constraints32,33.
As a compact, low-cost platform of high imaging performance, we expect that the wide adoption of the RA-WiFi mesoscope will benefit various studies of biodynamics in vivo, including neuroscience and immunology.
Methods
Optical setup
For the fluorescence imaging mode we used two LEDs, one at 470 nm (SOLIS-470C, Thorlabs) and one at 565 nm (SOLIS-565C, Thorlabs). Excitation light from the two LEDs is first combined via dichroic mirror DM1 (DMSP505L, Thorlabs) and then filtered using a multi-band excitation filter FEX (ZET405/488/561/640xv2, Chroma). To achieve structured-illumination-based optical sectioning, we modulated the excitation light with a DMD (DLP9500, Texas Instruments) conjugated to the sample plane. A total internal reflection prism was used to make the system compact. The modulated excitation beams are then relayed through two sets of relay lens pairs, one consisting of L1 (AC508-150-A, Thorlabs) and L2 (AC508-150-A), and the other one consisting of L3 (AC508-180-A, Thorlabs) and L4, which itself consists of four closely connected lenses of focal length 400 mm (AC508-400-A, Thorlabs) to minimize aberrations. The equivalent focal length of L4 is 102.8 mm. A manually tunable lens (ML-20-37, Optotune) was placed between L2 and L3 to compensate for the dioptres induced by imprecise instalment of the optical components. A multi-band dichroic mirror DM2 (ZT405/488/561/640rpcv2-UF1, Chroma) was used to reflect the excitation light into the ETL (EL-16-40-TC-VIS-5D-C, Optotune), which is conjugated to the objective’s back pupil. To achieve random-access scanning of the whole FOV, we introduced a two-axis galvo (6240H, Cambridge Technology) as close as possible to the ETL. The excitation light was then relayed to excite the sample through L5 and L6 (Supplementary Fig. 1) and then the objective (XLFLUOR4X/340, Olympus). The emitted fluorescence was epi-detected. After being collected by the same objective and reflected by the two-axis galvo, the fluorescence passes through the ETL and DM2, sequentially. We then relayed the fluorescence to the surface of the deformable mirror (DMP40/M-P01, Thorlabs) via a set of relay lens pairs, which consisted of SL1 (CLS-SL, Thorlabs) and SL2 (CLS-SL). The deformable mirror was used for aberration correction, and was conjugated to the back pupil plane of the objective. After passing through a multi-band emission filter FEM (ZET405/488/561/640mv2, Chroma), the aberration-corrected signal was imaged at the camera (Dhyana 400BSI V2, Tucsen) using tube lens TL (TTL180-A, Thorlabs).
Multiple imaging modes
By combining LEDs of two colours and multi-band filters, dual-colour fluorescence imaging is based on time-division multiplexing. For dark-field imaging, a white LED ring (MV-RL62X35W-A30-V, Microvision) with an illumination angle of 30° was attached to the objective. To achieve bright-field illumination, the distance between the ring LED and the sample was increased.
Random-access imaging scheme
To enable large-scale, high-resolution imaging using the RA-WiFi mesoscope, we choose a high space–bandwidth objective (XLFLUOR4X/340)34,35, which theoretically provides a lateral resolution of 1.1 μm for an emission wavelength at 0.52 μm and a FOV diameter of 5.5 mm. However, the theoretical resolution can be achieved only through Nyquist sampling. Considering a commercial sCMOS camera with representative parameters (Dhyana 400BSI V2: pixel size, 6.5 μm; pixel number, 2,048 × 2,048), the size of the camera pixel should be de-magnified to half of 1.1 μm at the sample plane. In such a case, the FOV diameter supported by the camera is 1.1/2 × 2,048 = 1.13 mm, which cannot cover the 5.5 mm FOV diameter supported by the objective. To bridge this gap, we proposed to randomly access a series of sub-FOVs, each with 1.1 μm resolution and a 1.13 mm FOV diameter, in sequence, to utilize the whole FOV. In practice, we set the system magnifications to produce a sub-FOV of 1.6 × 1.6 mm, except for imaging of the mouse spinal cords. To fully cover the width of the exposed mouse spinal cords, we adjusted the magnification of the lens pair SL2 and TL to obtain a 1.7 × 1.7 mm sub-FOV size (Fig. 4 and Supplementary Figs. 27–29).
In the fluorescence imaging mode, to direct signals from different sub-FOVs to project on to the same detector, we used a de-scan scheme (Fig. 1b; see also Supplementary Fig. 2). When the scan mirror is pointed at 45° with respect to the x axis (left panel of Supplementary Fig. 2), the scanner steers the excitation beam to the central area of the sample and reflects the collected fluorescence back to the camera. When the scanner is driven to induce an angular deflection (right panel of Supplementary Fig. 2), the excitation beam is steered to excite the edge area of the sample, and the excited fluorescence passes the scanner again, cancelling the angular deflection, and is collected by the camera.
In both the bright-field and dark-field imaging modes, we simultaneously illuminated the entire FOV but collected the sub-FOV images sequentially by steering the two-axis galvo.
Lower field angle design
We show the principle of the LFA configuration in Supplementary Fig. 3a. For general imaging applications, the object distance d (the distance between the sample plane and the principal plane of the objective) is equal to the objective’s nominal object distance (one focal length for the infinity-corrected objective), and the collected fluorescence exits the objective’s back pupil as a collimated beam and forms a diffraction-limited (DL) spot after the tube lens (Fig. 1c left; see also Supplementary Fig. 3b). In the DL configuration, the off-axis field of radius r exhibits the field angle α = arctan(r/d). In the LFA configuration, we move the sample plane to a position beyond the objective’s nominal object distance (that is, d + Δd), resulting the decreased field angle β = arctan(r/(d + Δd)), enabling the light emitted from a larger r to pass through the subsequent lenses and reach the camera, thereby extending the FOV. Meanwhile, the LFA configuration causes the fluorescence to exit the objective’s back pupil as a convergent beam (Fig. 1c middle and right; see also Supplementary Fig. 3b) and form as a spot that is larger than the DL spot, due to the loss of numerical aperture.
To validate the feasibility of the LFA configuration, we perform simulations using Zemax36. Since the Zemax model of the objective XLFLUOR4X/340 is unavailable, we model it using a perfect paraxial lens with a 45 mm focal length and a back aperture diameter of 25 mm. The tube lens, which consists of customized lenses (Supplementary Fig. 1), works as a 4f relay system together with the objective. When we set d to be equal to the nominal object distance of the objective (45 mm), owing to the limited diameter of the tube lens, beams emitted from the edge of the FOV are cut by the tube lens, leading to vignetting (left panel of Supplementary Fig. 3b). When we set d at 52 mm, beams emitted from the edge of the FOV can pass through the tube lens (right panel of Supplementary Fig. 3b). Simulation results of the fraction of unvignetted rays versus the FOV radius are shown in Supplementary Fig. 3c. When the FOV radius reaches 6.4 mm, ~50% of rays are cut by the tube lens in DL mode. However, no ray is cut in LFA mode. Meanwhile, as d increases, the effective numerical aperture decreases (Supplementary Fig. 3h).
System synchronization
In the RA-WiFi system, the small-step response time for the galvo scanner (6240H) is ~0.35 ms, the full-stroke response time for the deformable mirror (DMP40/M-P01) is ~0.5 ms and the response time for the DMD (DLP9500) is 0.06 ms; we thus set the switching time between the two ROIs to 1 ms in practical experiments.
The camera, DMD, LEDs, two-axis galvo, ETL and air-puff stimulus device of the RA-WiFi mesoscope are controlled using a data acquisition card (USB-6363, National Instruments). The deformable mirror is software-triggered and updated through a USB cable. We customized a LabVIEW program37 for hardware synchronization.
The synchronization diagram is shown in Supplementary Fig. 30. Briefly, one acquisition event of the camera consists of the exposure time and the readout time (minimal value: 15 ms). In single-colour random-access imaging mode, the camera obtains n DMD modulated images (n = 2 for the HiLo microscopy method and n = 3 for the refined structured illumination microscopy (SIM) method) to reconstruct an optical-sectioning image for one sub-FOV. The scanning controls, including the ETL, galvo and deformable mirror, update their configurations for the next sub-FOV during the readout time. Considering that the settling time of the scanning controls is less than 15 ms, devices can be settled down within the readout interval. In dual-colour random-access imaging mode, two LEDs alternately switch between on/off states for time-division multiplexing, and the camera takes 2n images (n = 2 for the HiLo method and n = 3 for the refined SIM method) for each sub-FOV. In the 1S2F mode, while the camera keeps on exposing, the scanning controls (the galvo and deformable mirror) alternate the configuration between two sub-FOVs, so n superimposed images of two sub-FOVs are obtained. During the alternating interval (1 ms) in 1S2F mode, the LED is switched to an off-state to avoid motion artefacts.
Field curvature correction
We use a liquid crystal display module as the calibration sample, and display a periodic pattern on it for field curvature calibration (Supplementary Fig. 3i). Before calibration, we use a dual-axis goniometer (GNL20/M, Thorlabs) and bubble level to avoid sample tilting. For each sub-FOV, several axial positions are scanned by applying a series of voltages to the ETL. At each axial position, the image sharpness (IS) is calculated as the metric function of field curvature correction. We define the image sharpness as IS = std(Iimage)/mean(Iimage), where std(Iimage) denotes the standard deviation of the intensity variations and mean(Iimage) indicates the averaged value of the image intensity. The sharpness values of each sub-FOV are Gaussian-fitted to find the optimum values. We show the experimentally calibrated field curvature correction in Supplementary Fig. 3j.
Sensorless adaptive optics
We apply sensorless AO to correct system aberrations, including the LFA configuration-induced aberrations. Specifically, a modally controlled deformable mirror (DMP40/M-P01) is conjugated to the objective’s back pupil, where aberrations can be represented as a linear combination of Zernike modes. We take a set of 12 low-order Zernike modes (excluding piston, tip and tilt) into consideration. We dilute fluorescent beads of 1.1 μm diameter (ThermoFisher) with 1% agarose as the measurement samples, and set the image sharpness (IS) as the metric function. Owing to the limited effective area of AO, we select the central 200 × 200 μm area of each sub-FOV for optimization. For each sub-FOV, we correct aberrations in a manner adapted from modal optimization38,39 (see Supplementary Note 1 for details).
Lateral resolution calibration
We dilute fluorescent beads of 1.1 μm diameter (ThermoFisher) in 1% agarose as samples. For each sub-FOV, we apply pre-calibrated field curvature correction and wavefront compensation on to the ETL and deformable mirror, respectively. To estimate the lateral resolution, we fit profiles of bead images with the two-dimensional Gaussian function and take the FWHM of the fitted results as the measured lateral resolution (Fm). The real lateral resolution (Fr) values are deconvoluted using \({F}_{{\mathrm{m}}}=\sqrt{F_{{\mathrm{r}}}^{2}+0.7464\times D_{{\mathrm{b}}}^{2}}\) (ref. 40) (see Supplementary Note 2), where Db is the diameter of the fluorescent beads. For each sub-FOV, several beads are randomly selected for lateral resolution calibration.
Optical sectioning via structured illumination and computational reconstruction
For the in vitro experiments, we achieve optical sectioning using the HiLo method41 in which two raw images are recorded to reconstruct one optical-sectioning image, decreasing the imaging speed by half. We thus use the refined SIM method with interleaved reconstruction to achieve optical sectioning in the in vivo experiments42,43. Briefly, three raw images (I1, I2 and I3), each laterally shifted by a third of the grid period, are acquired. The optical-sectioning images are computationally reconstructed via IRSIM = HP(IU) + ηLP(ICSIM), where HP(IU) denotes the high-pass Gaussian filtering of the uniformly illuminated image IU = [I1 + I2 + I3]/3, LP(ICSIM) indicates the complementary low-pass Gaussian filtering of conventional SIM image \({I}_{{{\mathrm{CSIM}}}}=\)\(\sqrt{{\left({I}_{1}-{I}_{2}\right)}^{2}+{\left({I}_{1}-{I}_{3}\right)}^{2}+{\left({I}_{2}-{I}_{3}\right)}^{2}}\) and η is a scaling factor to ensure continuity at the crossover frequency in the Fourier domain42. To fill in the missing temporary information among adjacent reconstructed frames, we utilize the interleaved refined reconstruction method43 in which we slide the time windows to pick up raw images for reconstruction.
To evaluate the optical-sectioning capabilities, we apply a thin layer of fluorescent paint (TS-36, Tamiya) on to glass coverslips via spin coating and use this layer as a test sample. For each sub-FOV, we first calibrate the axial position of the fluorescent layer (the position when the uneven grains are sharply focused) and set the focal plane 100 µm above the layer, then keep lifting the sample in 5 µm steps by moving the sample stage (MT3/M-Z8, Thorlabs) until the focal plane is 200 µm below the layer. For each axial plane, we record three structured illuminated images, each with a phase shift of 2π/3. For each axial plane, we reconstruct the optical-sectioning image and use the mean intensity to indicate the axial response. We show the experimental evaluation of the optical-sectioning capabilities in Supplementary Fig. 4.
Multi-plane imaging of curved surfaces in vivo
To perform the multi-plane imaging of mouse brain samples with intact curved skulls and mouse spinal cords in vivo, we scan the axial positions by applying a series of voltages to the ETL and determine the optimal focusing planes of each sub-FOV before imaging for biodynamics. By performing Gaussian fitting of the image sharpness across various depths, we obtain the best focused axial position of each sub-FOV. When the sharpness fitting fails to focus on the biological structures (mostly caused by intense artefacts that remain on the sample surface, such as glue used in fixation), we manually adjust the axial positions of the failed sub-FOVs.
Tilt calibration and tilted sample imaging
The RA-WiFi mesoscope can calibrate sample tilting via three-times measurements. We select three non-collinear sub-FOVs and scan several axial positions to search for their optimal focusing depths by applying a series of voltages to the ETL at each sub-FOV. By performing Gaussian fitting of the image sharpness across various depths, we obtain the optimal ETL voltage to reach maximum sharpness. In following this procedure, we can obtain three-dimensional coordinates for the measured sub-FOVs: (xi, yi, zi), where i = 1, 2, 3. The equation of the tilted plane can be fitted by substituting (xi, yi, zi) into Ax + By + Cz + D = 0, and the ETL voltages for tilted sample imaging can be calculated by adding the tilt correction voltages to the field curvature correction voltages.
1S2F scheme and signal de-mixing
In conventional random-access mode, the overall frame rate (FRRA) of the RA-WiFi mesoscope is limited by the number of targeted sub-FOVs (m), the exposure time for each sub-FOV (texpo) and the minimal readout time (tr = 15 ms) of the camera, as FRRA = 1/(m × (tr + texpo)). When texpo is tens to several tens of milliseconds, the readout time becomes the bottleneck of FRRA. To address this problem, we propose the 1S2F scheme. In the 1S2F scheme, as the two-axis galvo and deformable mirror alternate their settings from one sub-FOV to the other sub-FOV, images of two sub-FOVs are recorded in one camera-exposure event, and the resulting image is equivalent to the superposition of the images in two sub-FOVs. The frame rate of the 1S2F scheme is FR1S2F = 1/(tr + 2 × texpo + ta), where ta is the minimal alternating time (in milliseconds, typically 1 ms in our current system for two sub-FOVs with a small focal depth difference) for the scanning controls. Compared with the conventional random-access mode, the imaging speed increases by
To de-mix the signals recorded in the 1S2F scheme, we first acquire previous information (for example, the neuron spatial distribution or the vessel structures) by carrying out conventional random-access imaging of selected ROIs. Then we record images in the 1S2F scheme and compare the 1S2F images with the previous information to attribute the signals to their corresponding sub-FOVs.
Image stitching
We set the overlap between adjacent sub-FOVs to be 5%. Before seamless stitching, we perform vignetting correction. Specifically, for each sub-FOV, we record a series of images with different contents (around ten tiles) and then apply average intensity projection on them to obtain the initial vignetting profile. To remove the sample-induced foreground information while keeping the vignetting profile, we further apply a border-limited mean filter44 to the initial vignetting profile. The vignetting-free image is then obtained by dividing the raw image by the vignetting profile. The whole FOV image is then stitched using Stitching plugin of ImageJ45.
Animal experiments
All procedures involving animals are approved by the Animal Care and Use Committees of Tsinghua University. Mice older than eight weeks are used unless specifically noted. See Supplementary Note 3 for additional details of the surgical preparations.
For cortex-wide calcium imaging under physiological conditions, we used GCaMP6f-expressing Ai148D/Rasgrf mice (Jackson Laboratory: strain numbers 030328 and 022864). To induce GCaMP expression in Ai148D/Rasgrf mice, trimethoprim (Sigma) was injected at 0.5 mg per g (body weight)46. After craniotomy, we performed acute imaging in lightly anaesthetized mice (using 0.5% isoflurane) to monitor spontaneous neuronal activities and in awake mice for functional imaging under sensory stimulation. For cortex-wide calcium imaging under pathological conditions, we use both GCaMP6f-expressing Ai148D/Rasgrf mice and GCaMP6s-expressing Ai162/PV mice (Jackson Laboratory: strain numbers 031562 and 017320). After craniotomy and two weeks’ recovery, KA (Sigma) was injected intraperitoneally at 10 mg per kg (body weight) to induce epilepsy18. Twenty minutes after the KA injection, we performed imaging in awake mice.
For the imaging of mouse pups with intact skulls, we used GCaMP6s-expressing Ai162/Emx1 mice (Jackson Laboratory: strain numbers 031562 and 005628) aged at P4–P5 and performed imaging in awake mice.
For the dual-colour imaging of neurovascular coupling, we uses Thy1-jRGECO1 mice (Jackson Laboratory: strain number 030526). After craniotomy and two weeks’ recovery, we injected FITC (Mw 200,000; Sigma Aldrich) into the mouse orbital vein (2% w/v in saline, 200 mg per kg (body weight)) under anaesthetization (~2.5% isoflurane), and performed imaging in awake mice.
For the imaging of dye perfusion in the mouse spinal cord, after laminectomy on C57BL/6 mice, several seconds after we began the acute imaging, FITC fluorescent dye (2% w/v; FD2000S, Sigma Aldrich) was administered via the orbital route simultaneously as the imaging was going on. For the imaging of neutrophil trafficking, retro-orbital injection of Ly-6G (1% w/v; ANTI-MO LY-6G RB6-8C5 PE, ThermoFisher) to C57BL/6 mice was performed 1–2 h after laminectomy, and the in vivo imaging is performed 10 min after injection. For calcium imaging in the mouse spinal cord, we used GCaMP6f-expressing Ai148D/Camk2a-Cre (Jackson Laboratory: strain numbers 030328 and 005359). After laminectomy, we fixed a rectangular coverglass on the exposed spinal cord, and after recovery for three days we performed imaging in awake mice.
Data processing
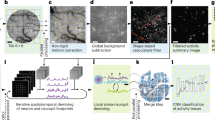
For data from in vivo experiments, we used the non-rigid version of the NoRMCorre algorithm for motion correction47.
For cortex-wide functional imaging under physiological conditions, we detected the neurons using the CNMF-e approach48.
For functional network building (Fig. 2c and Supplementary Figs. 14 and 15h), we first calculated the pairwise distances between the mass centres of neural contours, then calculated the pairwise CCs between the neural calcium signals. We regarded neurons with CC values above 0.4 as nodes and mapped pairwise links between the nodes.
To evaluate the contrast enhancement of structured-illumination-based optical sectioning (Fig. 2i and Supplementary Fig. 19c), we defined the contrast as C =std(ΔF/F)/mean(ΔF/F), where std(ΔF/F) denotes the standard deviation of ΔF/F, and mean(ΔF/F) denotes the average value of ΔF/F.
For cortex-wide functional imaging under pathological conditions, we segmented neuronal ROIs corresponding to visually identifiable cell bodies using a semiautomated algorithm49. For the spatiotemporal maps of successive neuron recruitment (Fig. 3a,f and Supplementary Figs. 23a and 26a), we selected a seizure event that involved most neurons within our observation area and then calculated the first discrete derivative (slope) of each neuronal activity trace. We regarded the maximal point of the slope as the onset moment for cell recruitment to the seizure event16 and sorted the activity of the neurons based on the order of onset. To depict the propagation patterns of seizure (Fig. 3d,e,i,j and Supplementary Figs. 24 and 25), several directional lines were drawn from each epileptic focus, and the intensity changes of the pixels along these lines were self-normalized to facilitate comparison. To measure the lasting time of the hyperactive state pixelwise along these paths, we defined the lasting time of a pixel as the width between time points where the intensity reached 10% of the maximal intensity.
For the analysis of neurovascular coupling (Figs. 3c,h and 5j and Supplementary Figs. 9, 10, 22 and 26), we extracted the diameter variations of blood vessels using the DiameterJ plugin of ImageJ50.
To compare the images acquired with and without multi-plane imaging, we performed Fourier analysis (Fig. 4c), following the method in ref. 51. The stitched whole FOV images were first transformed to Fourier space, and the background noise power, which is identified as the median value of the spectra outside the diffraction-limited frequency, is subtracted from the Fourier spectra. Then, we found the maximum frequency with a 0.02% level of the zero frequency power and denoted its radial distance as zero frequency.
For the trajectory analysis of neutrophil trafficking, we first subtracted the temporally averaged background from the raw images to enhance the contrast. Then we extracted the neutrophil traces manually using the MTrackJ plugin of ImageJ52. The trafficking velocity was calculated using the time interval and the straight line distance between the neutrophil positions in two successive frames.
For the analysis of dye perfusion in the blood vessels (Supplementary Fig. 27c), after motion registration we extracted all pixels within the spinal cord window via intensity thresholding of the vessel images during a stable stage (35 s after dye injection). For each pixel, the fluorescence signal was first normalized to the intensity in the stable stage, and then the signal was filtered using a Butterworth low-pass filter. The time point when the smoothed signal increased to 5% of the stable level was defined as the rising start time.
For calcium signal analysis in the mouse spinal cords (Supplementary Fig. 29), we followed the method based on gridded ROIs in ref. 19. Briefly, we first used intensity thresholding and morphology operation to filter out the pixels on or near the vessels. Then, the non-vessel area was divided into gridded ROIs of ~13 × 13 μm, and ΔF/F (∆F/F = (F−F0)/F0, F0 is the minimum value in recording) was calculated for each ROI. For clustering analysis, first, the Pearson CC matrix of the gridded ROIs was calculated. Each ROI has a feature vector of its CCs with the other ROIs, and the Manhattan distance (L1 norm) between the feature vector and the zero vector was calculated. We applied hierarchical clustering according to the L1 feature distance, with the inter-cluster threshold set as 50% of the maximum feature distance.
Network architecture and training details
The network architecture of self-supervised restoration uses contrastive unpaired translation, which is reported to have a superior performance on unpaired image-to-image translation33. In general, the network is composed of the ResNet-based generator with nine residual blocks53, the PatchGAN discriminator54, PatchNCE loss31 and Least Squares GAN loss55. The Adam optimizer was used for network training, with a learning rate of 0.002 (ref. 56). Two graphics processing units (Nvidia GeForce RTX 1080 Ti, 12 GB memory) were used to accelerate the training and testing process.
The datasets used to train the network were generated via single large-FOV RA-WiFi imaging according to the aberration intensity distribution prior. Owing to previous knowledge that aberrations in peripheral regions are more severe than those in central regions (Supplementary Fig. 31a), we can naturally obtain unpaired high and low aberration data for training from different spatial locations of the same FOV (Supplementary Fig. 31b). In addition, owing to the large FOV of RA-WiFi mesoscopy, only single-shot imaging can meet the demand size for training data. Thus we can use large-FOV imaging directly for post-hoc aberration restoration, without having to build additional optical paths or simulations to prepare the training data.
Data availability
The raw and analysed datasets generated during the current study are too large to deposit in a public repository but they are available from the corresponding author upon reasonable request. The related data files that support our results are available via Figshare at https://doi.org/10.6084/m9.figshare.24557326 (ref. 57).
Code availability
The codes that support the findings of this study are available from the corresponding author upon reasonable request.
References
Tyson, A. L. & Margrie, T. W. Mesoscale microscopy and image analysis tools for understanding the brain. Prog. Biophys. Mol. Biol. 168, 81–93 (2022).
Park, J., Brady, D., Zheng, G., Tian, L. & Gao, L. Review of bio-optical imaging systems with a high space–bandwidth product. Adv. Photonics 3, 044001 (2021).
Potsaid, B., Bellouard, Y. & Wen, J. T. Adaptive Scanning Optical Microscope (ASOM): a multidisciplinary optical microscope design for large field of view and high resolution imaging. Opt. Express 13, 6504–6518 (2005).
Sofroniew, N. J., Flickinger, D., King, J. & Svoboda, K. A large field of view two-photon mesoscope with subcellular resolution for in vivo imaging. eLife 5, e14472 (2016).
Fan, J. et al. Video-rate imaging of biological dynamics at centimetre scale and micrometre resolution. Nat. Photonics 13, 809–816 (2019).
Zhou, K. C. et al. Parallelized computational 3D video microscopy of freely moving organisms at multiple gigapixels per second. Nat. Photonics 17, 442–450 (2023).
McConnell, G. et al. A novel optical microscope for imaging large embryos and tissue volumes with sub-cellular resolution throughout. eLife 5, e18659 (2016).
Yu, C. H., Stirman, J. N., Yu, Y., Hira, R. & Smith, S. L. Diesel2p mesoscope with dual independent scan engines for flexible capture of dynamics in distributed neural circuitry. Nat. Commun. 12, 6639 (2021).
Ota, K. et al. Fast, cell-resolution, contiguous-wide two-photon imaging to reveal functional network architectures across multi-modal cortical areas. Neuron 109, 1810–1824 (2021).
Janiak, F. K. et al. Non-telecentric two-photon microscopy for 3D random access mesoscale imaging. Nat. Commun. 13, 544 (2022).
Ren, C. & Komiyama, T. Characterizing cortex-wide dynamics with wide-field calcium imaging. J. Neurosci. 41, 4160–4168 (2021).
Urai, A. E., Doiron, B., Leifer, A. M. & Churchland, A. K. Large-scale neural recordings call for new insights to link brain and behavior. Nat. Neurosci. 25, 11–19 (2022).
Hauser, W. A. Seizure disorders: the changes with age. Epilepsia 33, 6–14 (1992).
Somarowthu, A., Goff, K. M. & Goldberg, E. M. Two-photon calcium imaging of seizures in awake, head-fixed mice. Cell Calcium 96, 102380 (2021).
Tran, C. H. et al. Interneuron desynchronization precedes seizures in a mouse model of Dravet syndrome. J. Neurosci. 40, 2764–2775 (2020).
Wenzel, M., Hamm, J. P., Peterka, D. S. & Yuste, R. Acute focal seizures start as local synchronizations of neuronal ensembles. J. Neurosci. 39, 8562–8575 (2019).
Rosch, R. E., Hunter, P. R., Baldeweg, T., Friston, K. J. & Meyer, M. P. Calcium imaging and dynamic causal modelling reveal brain-wide changes in effective connectivity and synaptic dynamics during epileptic seizures. PLoS Comput. Biol. 14, e1006375 (2018).
Sperk, G. Kainic acid seizures in the rat. Prog. Neurobiol. 42, 1–32 (1994).
Shekhtmeyster, P. et al. Trans-segmental imaging in the spinal cord of behaving mice. Nat. Biotechnol. 41, 1729–1733 (2023).
Xue, Y., Davison, I. G., Boas, D. A. & Tian, L. Single-shot 3D wide-field fluorescence imaging with a Computational Miniature Mesoscope. Sci. Adv. 6, eabb7508 (2020).
Hoffmann, A. et al. High and low molecular weight fluorescein isothiocyanate (FITC)–dextrans to assess blood-brain barrier disruption: technical considerations. Transl. Stroke Res. 2, 106–111 (2011).
Iwai, D., Izawa, H., Kashima, K., Ueda, T. & Sato, K. Speeded-up focus control of electrically tunable lens by sparse optimization. Sci. Rep. 9, 12365 (2019).
Mac, K. D. et al. Fast volumetric imaging with line-scan confocal microscopy by electrically tunable lens at resonant frequency. Opt. Express 30, 19152–19164 (2022).
Yuan, X., Brady, D. J. & Katsaggelos, A. K. Snapshot compressive imaging: theory, algorithms, and applications. IEEE Signal Process. Mag. 38, 65–88 (2021).
Shi, Z., Xu, X., Liu, X., Chen, J. & Yang, M.-H. Video frame interpolation transformer. In Proc. IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) 17482–17491 (IEEE, 2022)
Kalluri, T., Pathak, D., Chandraker, M. & Tran, D. FLAVR: flow-agnostic video representations for fast frame interpolation. In Proc. IEEE/CVF Winter Conference on Applications of Computer Vision (WACV) 2071–2082 (IEEE, 2023)
Mahecic, D. et al. Event-driven acquisition for content-enriched microscopy. Nat. Methods 19, 1262–1267 (2022).
Liu, M.-Y., Breuel, T. & Kautz, J. Unsupervised image-to-image translation networks. In Advances in Neural Information Processing Systems 30 (eds von Luxburg, U. et al.) 701–709 (Neural Information Processing Systems Foundation, 2017).
Liang, W. et al. Advances, challenges and opportunities in creating data for trustworthy AI. Nat. Mach. Intell. 4, 669–677 (2022).
Creswell, A. et al. Generative adversarial networks: an overview. IEEE Signal Process. Mag. 35, 53–65 (2018).
Park, T., Efros, A. A., Zhang, R. & Zhu, J.-Y. Contrastive learning for unpaired image-to-image translation. In European Conference on Computer Vision 2020 (ECCV 2020) (eds Vedaldi, A. et al.) 319–345 (Springer, 2020)
Weigert, M. et al. Content-aware image restoration: pushing the limits of fluorescence microscopy. Nat. Methods 15, 1090–1097 (2018).
Belthangady, C. & Royer, L. A. Applications, promises, and pitfalls of deep learning for fluorescence image reconstruction. Nat. Methods 16, 1215–1225 (2019).
Bumstead, J. R. Designing a large field-of-view two-photon microscope using optical invariant analysis. Neurophotonics 5, 025001 (2018).
Tsai, P. S. et al. Ultra-large field-of-view two-photon microscopy. Opt. Express 23, 13833–13847 (2015).
OpticStudio v.19.4 (Zemax, 2024).
LabVIEW (National Instruments, 2019).
Verstraete, H. R., Wahls, S., Kalkman, J. & Verhaegen, M. Model-based sensor-less wavefront aberration correction in optical coherence tomography. Opt. Lett. 40, 5722–5725 (2015).
Bonora, S. & Zawadzki, R. J. Wavefront sensorless modal deformable mirror correction in adaptive optics: optical coherence tomography. Opt. Lett. 38, 4801–4804 (2013).
Theer, P., Mongis, C. & Knop, M. PSFj: know your fluorescence microscope. Nat. Methods 11, 981–982 (2014).
Lim, D., Ford, T. N., Chu, K. K. & Mertz, J. Optically sectioned in vivo imaging with speckle illumination HiLo microscopy. J. Biomed. Opt. 16, 016014 (2011).
Li, Z. et al. Fast widefield imaging of neuronal structure and function with optical sectioning in vivo. Sci. Adv. 6, eaaz3870 (2020).
Shi, R., Li, Y. & Kong, L. High-speed volumetric imaging in vivo based on structured illumination microscopy with interleaved reconstruction. J. Biophotonics 14, e202000513 (2021).
Johnson, K. A. & Hagen, G. M. Artifact-free whole-slide imaging with structured illumination microscopy and Bayesian image reconstruction. Gigascience 9, giaa035 (2020).
Preibisch, S., Saalfeld, S. & Tomancak, P. Globally optimal stitching of tiled 3D microscopic image acquisitions. Bioinformatics 25, 1463–1465 (2009).
Harris, J. A. et al. Anatomical characterization of Cre driver mice for neural circuit mapping and manipulation. Front. Neural Circuits 8, 76 (2014).
Pnevmatikakis, E. A. & Giovannucci, A. NoRMCorre: an online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94 (2017).
Zhou, P. et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. eLife 7, e28728 (2018).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Fischer, M. J., Uchida, S. & Messlinger, K. Measurement of meningeal blood vessel diameter in vivo with a plug-in for ImageJ. Microvasc. Res. 80, 258–266 (2010).
Royer, L. A. et al. Adaptive light-sheet microscopy for long-term, high-resolution imaging in living organisms. Nat. Biotechnol. 34, 1267–1278 (2016).
Meijering, E., Dzyubachyk, O. & Smal, I. Methods for cell and particle tracking. Methods Enzymol. 504, 183–200 (2012).
Johnson, J., Alahi, A. & Fei-Fei, L. Perceptual losses for real-time style transfer and super-resolution. In European Conference on Computer Vision 2016 (ECCV 2016) (eds Leibe, B. et al.) 694–711 (Springer, 2016).
Isola, P., Zhu, J.-Y., Zhou, T. & Efros, A. A. Image-to-image translation with conditional adversarial networks. In 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 5967–5976 (IEEE, 2017).
Mao, X., et al. Least squares generative adversarial networks. In 2017 IEEE International Conference on Computer Vision (ICCV) 2813–2821 (IEEE, 2017).
Kingma, D. P. & Ba, J. L. Adam: A method for stochastic gradient descent. In 2015 International Conference on Learning Representations (ICLR) (ICLR Press 2015).
Shi, R. et al. Random-access wide-field mesoscopy for centimetre-scale imaging of biodynamics with subcellular resolution. Figshare https://doi.org/10.6084/m9.figshare.24557326 (2024).
Acknowledgements
This project is supported by the STI2030-Major Projects (number 2022ZD0212000, to L.K.), National Natural Science Foundation of China (NSFC) (numbers 61831014 and 32021002, both to L.K.) and a ‘Bio-Brain+X’ Advanced Imaging Instrument Development Seed Grant (to L.K.).
Author information
Authors and Affiliations
Contributions
L.K. conceived the idea and supervised the project. R.S. designed and manufactured the system, with X.C. providing assistance. L.Z. and P.T. provided suggestions on the biological experimental design. R.S., X.C. and J.D. performed biological experiments with the assistance of K.F. (sample preparation and craniotomy). R.S. and X.C. analysed the experimental data, with suggestions from F.Z. The deep-learning framework was developed by J.L. The manuscript was prepared by R.S., X.C. and L.K., with inputs from all co-authors.
Corresponding authors
Ethics declarations
Competing interests
L.K. and R.S. have granted patents on the presented methods. The other authors declare no competing interests.
Peer review
Peer review information
Nature Photonics thanks Roarke Horstmeyer and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary Information
Supplementary Notes 1–3, Figs. 1–31 and Table 1.
Supplementary Video 1
Cortex-wide functional imaging of spontaneous neural activity in mice in vivo. The imaging FOV is 6.4 × 6.4 mm, the frame rate is 1.67 Hz and the playback speed is ×8. The calcium activity of neurons (red dots) is superimposed on the minimum intensity projection background (grey).
Supplementary Video 2
Cortex-wide functional imaging of stimuli-induced neural activity in mice in vivo. The imaging FOV is 6.4 × 6.4 mm, the frame rate is 1 Hz and the playback speed is ×8. The calcium activity of neurons (red dots) is superimposed on the minimum intensity projection background (grey).
Supplementary Video 3
Random-access imaging of three arbitrarily assigned regions of stimuli-induced neural activity in mice in vivo. The recording area includes three sub-FOVs, each of 1.6 × 1.6 mm. The frame rate is 5.12 Hz and the playback speed is ×2. The calcium activity of neurons (red dots) is superimposed on the minimum intensity projection background (grey).
Supplementary Video 4
Simultaneous functional and structural imaging of neurovascular coupling in seizures of Ai148D/Rasgrf mice in vivo. The imaging FOV is 4.8 × 6.4 mm, the frame rate is 1.67 Hz and the playback speed is ×12.
Supplementary Video 5
Simultaneous functional and structural imaging of neurovascular coupling in seizures of Ai162/PV mice in vivo. The imaging FOV is 4.8 × 4.8 mm, the frame rate is 2.8 Hz and the playback speed is ×9.
Supplementary Video 6
Calcium imaging of seizures spreading in the cortex of Ai148D/Rasgrf mice in vivo. The imaging FOV is 4.8 × 3.2 mm, the frame rate is 3.3 Hz and the playback speed is ×6. The fluorescence intensity variation (red) is superimposed on the minimum intensity projection background (grey).
Supplementary Video 7
Calcium imaging of seizures spreading in the cortex of Ai162/PV mice in vivo. The imaging FOV is 4.8 × 4.8 mm, the frame rate is 2.8 Hz and the playback speed is ×5. The fluorescence intensity variation (red) is superimposed on the minimum intensity projection background (grey).
Supplementary Video 8
High-speed imaging of dye perfusion in the blood vessels of C57BL/6 mouse spinal cord in vivo. The imaging FOV is 1.7 × 6.5 mm, the frame rate is 9.6 Hz and the playback speed is ×1.
Supplementary Video 9
Imaging of neutrophil trafficking along the blood vessels in C57BL/6 mouse spinal cords in vivo. The imaging FOV is 1.7 × 6.5 mm, the frame rate is 3.3 Hz and the playback speed is ×3. Traces in the most recent 1.8 s of each time point are displayed and labelled using random colours.
Supplementary Video 10
Calcium imaging of spontaneous neural activity in Ai148D/Camk2a-Cre mouse spinal cord in vivo. The imaging FOV is 1.7 × 4.9 mm, the frame rate is 6.7 Hz and the playback speed is ×2. The fluorescence intensity variation (red) is superimposed on the minimum intensity projection background (grey).
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shi, R., Chen, X., Deng, J. et al. Random-access wide-field mesoscopy for centimetre-scale imaging of biodynamics with subcellular resolution. Nat. Photon. (2024). https://doi.org/10.1038/s41566-024-01422-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41566-024-01422-1