Abstract
Epithelial-to-mesenchymal transition (EMT) is fundamental to both normal tissue development and cancer progression. We hypothesized that EMT plasticity defines a range of metabolic phenotypes and that individual breast epithelial metabolic phenotypes are likely to fall within this phenotypic landscape. To determine EMT metabolic phenotypes, the metabolism of EMT was described within genome-scale metabolic models (GSMMs) using either transcriptomic or proteomic data from the breast epithelial EMT cell culture model D492. The ability of the different data types to describe breast epithelial metabolism was assessed using constraint-based modeling which was subsequently verified using 13C isotope tracer analysis. The application of proteomic data to GSMMs provided relatively higher accuracy in flux predictions compared to the transcriptomic data. Furthermore, the proteomic GSMMs predicted altered cholesterol metabolism and increased dependency on argininosuccinate lyase (ASL) following EMT which were confirmed in vitro using drug assays and siRNA knockdown experiments. The successful verification of the proteomic GSMMs afforded iBreast2886, a breast GSMM that encompasses the metabolic plasticity of EMT as defined by the D492 EMT cell culture model. Analysis of breast tumor proteomic data using iBreast2886 identified vulnerabilities within arginine metabolism that allowed prognostic discrimination of breast cancer patients on a subtype-specific level. Taken together, we demonstrate that the metabolic reconstruction iBreast2886 formalizes the metabolism of breast epithelial cell development and can be utilized as a tool for the functional interpretation of high throughput clinical data.
Similar content being viewed by others
Introduction
Roughly 90% of all cancer-related deaths are believed to be caused by secondary metastatic tumors1. Multiple enzymes have been identified that support cancer cell dissemination in breast cancer through alterations of core metabolic pathways. These include the glycolytic enzymes HK1 and PKM22,3, IDH1 involved in the tricarboxylic acid (TCA) cycle4, ACLY in fatty acid synthesis5, and PRODH from proline synthesis6. Definitive metabolic patterns that differentiate between invasive and non-invasive cancer cells however remain elusive7.
One way that epithelial cells gain invasive properties is through the developmental process known as epithelial-to-mesenchymal transition (EMT). When localized breast cancer epithelial cells go through EMT, they gain invasive and apoptosis-resistant properties that contribute to their ability to migrate through the extracellular matrix and form secondary tumors through mesenchymal-to-epithelial transition (MET)8,9,10. Metabolic alterations are believed to be a hallmark of cancer and tumor progression11 and thus, an overall understanding of the metabolic changes that accompany EMT and MET in breast tissue may help to recognize potential biomarkers and drug targets associated with cancer progression.
Genome-scale metabolic models (GSMMs) have been successfully used to analyze and interpret changes to cancer metabolism based upon high-throughput datasets12,13,14. GSMM-based studies have revealed significant alterations in the reducing potential during breast tumor development where NADPH is increasingly directed towards reactive oxygen species (ROS) defenses15. Furthermore, the predicted metabolic variability between patients has been utilized successfully for their prognosis14. These studies14,15 were based on transcriptomic or proteomic data obtained from the cell lines or tumors of interest but lacked direct measurements of uptake/secretion rates that constrain metabolic flux as these measurements are challenging to obtain in a clinical setting. Directly incorporating metabolic measurements is expected to provide more accurate predictions than clinical breast cancer data alone.
We hypothesize that GSMMs representing the metabolic plasticity of EMT may help define the metabolism of breast tissue and contribute to the identification of metabolic vulnerabilities for breast cancer diagnostic or therapeutic purposes. The epithelial-derived D492 cell EMT model is comprised of two cell lines (D492 and D492M) that allow metabolic differences that occur following spontaneous EMT in cell culture to be investigated16. Similar cell models previously used to study EMT include HMLE and the PMC42 EMT cell models17,18,19.
In order to describe the metabolic plasticity of EMT we recently reported the metabolism of D492 and its mesenchymal-like counterpart D492M by integrated analyses of extracellular metabolomic- and transcriptomic data within tailored GSMMs. The metabolic alterations that occur following EMT in D49216 mirrored results from a comprehensive analysis of EMT metabolism20 and anchorage-independent growth21. A decrease in glycolysis and changes to mitochondrial oxidation of amino acids, specifically glutamine, threonine, arginine and lysine were observed. Those analyses were limited to transcriptomic and extracellular metabolomics data prompting the question of how proteomic data would alter the predictions of D492 metabolic network activity given the nonlinear relationship of transcription and translation22,23.
Here, we extend the D492 EMT GSMM, now termed iBreast2886, to include differences in protein levels, further formulating the metabolism of EMT and investigate the models ability to describe breast tissue metabolism. In order to capture the intracellular metabotypes that accompany EMT in D492 and identify biomarkers that discriminate between the two phenotypes, we used constraint-based modeling and comparative metabolic analysis. In order to reconcilidate the predicted differences in metabolic phenotypes based on the different data types, we carried out enzyme inhibitor assays, 1-13C-glutamine tracer analyses, and siRNA knockdown experiments in vitro to determine the actual phenotypes D492 and D492M cells. Finally, we demonstrate how iBreast2886 can be used as a tool for functional interpretation of tumor gene expression data from breast cancer patients.
Results
Direct comparison of different data types reveals their low overlap
In order to determine the consistency of the three different types of data used in this study (microarray, RNA sequencing (RNA-seq) and proteomic) for D492 epithelial cells and D492M mesenchymal cells, we compared the three data types by calculating the Spearman correlation of the log-fold differences between D492M and D492 (Fig. 1a).
The correlation between RNA-seq and proteomic data was the highest (ρ = 0.46) and the correlation between the two gene expression methods was lowest (ρ = 0.28). By comparing only the metabolic identifiers, the correlation between the dataset did not change (Fig. 1b).
To compare the datasets on a more functional metabolic level, we investigated and compared their ability to infer metabolic activity of D492 and D492M using constraint-based metabolic modeling24. In order to achieve this, we used the different datasets as constraints on our previous reconstruction of breast metabolism, which we refer to hereafter as iBreast2886.
True metabolic flux is reflected in cell-specific metabolic networks from proteomic data but not other data types
For the comparative metabolic analysis, we constructed GSMMs based on RNA-seq and proteomic data from the epithelial D492 and mesenchymal D492M and compared these to microarray-based GSMMs built previously16 and iBreast2886 GSMMs where only the extracellular constraints were applied. Henceforth, these will be referred to as the RNA-seq GSMMs, protein GSMMs, microarray GSMMs, and media GSMMs.
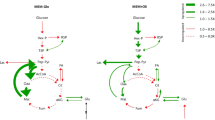
In order to compare the EPI and MES models in all pairs of GSMMs, representative flux values (flux profiles) for all reactions that obey the GSMM steady-state assumptions for all models were obtained through random sampling of the solution space25, where the median values for reactions were used to represent their activity. The relative differences between EPI and MES in all four GSMM pairs are summarized in Fig. 2a–d. Hierarchical clustering of the GSMMs flux profiles revealed highest similarity between the RNA-seq- and proteomic-constrained models on a phenotype-specific level (Supplementary figure 1). Upon closer inspection, it was clear that reaction similarity was different in the various subcellular compartments. Specifically, the flux similarity of the RNA-seq- and proteomic-constrained models was compartment specific, where the endoplasmic reticulum (ER) had the highest correlation of EMT-linked differences in reaction activity, followed by the cytosol and mitochondria (Supplementary table 1).
a Only extracellular constraints, (b) Microarray, (c) RNA-seq and (d) Proteomic data. The pathways shown are glycolysis, TCA cycle, and pentose-phosphate pathway. Red represents higher activity in MES whereas blue represents higher activity in EPI, represented by log-fold differences in median activity from random sampling of the models.
As the ground truth for the comparative analysis of pathway activity within our GSMMs, we used isotope labeling patterns from 1-13C-labeled glutamine experiments. This tracer is capable of quantifying the contribution of glutamine, one of two major carbon sources of D492 and D492M16, to citrate, malate, and aspartate through reductive carboxylation (Fig. 3a) and to the synthesis of proline and glutathione. The contributions from glutamine to the aforementioned metabolites are not whole metabolic pathways but subsets of reductive glutaminolysis and will be referred to as metabolic routes. Some of these metabolic routes occur in more than one cellular compartment. Reductive glutaminolysis is therefore a good representation of the compartment-based complexity of eukaryotic cellular metabolism.
a A carbon tracing map of 1-13C-labeled glutamine describing the flow and fate of labeled carbons in the glutamine carbon skeleton. Metabolites in bold are the end metabolites within the pathways we quantified. Red circles represent the 13C- carbon isotopes. The mitochondria is indicated by shaded grey. b Density plots of the calculated metabolic route activitiy (MRA) of five different routes of reductive glutamine metabolism from the total random sampling matrix (n = 5800 flux vectors) for all GSMMs. The blue distributions represent MRA within the epithelial GSMMs whereas red represents MRA within the mesenchymal GSMMs. The dashed line represents the median MRA value. Higher (i.e. more positive) values represent more active routes. All distributions were significantly different in (b) (p < 0.001) based on a Kolmogorov–Smirnov test. c Measured total contribution (TC) from 1-13C-glutamine to selected metabolites (after 6 h of cell culture) which represent the same metabolic routes as in (b). Results in (c) are shown as mean + SEM from three experiments (shown with dots). Student’s t-test was used to estimate significance.
It is challenging to infer metabolic pathway activity by observing multiple, individual reactions (c.f Fig. 2). Therefore, we utilized an activity measure that quantifies metabolic route activity in compartmentalized GSMMs based on random sampling results. From the metabolic route activity calculations, we observed that the different omics-constrained GSMMs had different predictions of the production of metabolites derived from glutamine (Fig. 3b). According to the 1-13C-labeled glutamine results, there was relatively higher citrate derived from glutamine in D492M than D492, indicating increased flow through reductive carboxylation of glutamine and/or the decreased condensation of oxaloacetate and acetyl-CoA (Fig. 3a). The proteomic GSMMs were the only ones to predict both routes correctly (Fig. 3c and Supplementary figure 2) along with the other four routes that were investigated. The microarray-constrained GSMMs correctly predicted the relative difference in metabolic route activity for only two routes, the RNA-seq GSMMs for three routes, and the GSMMs constrained only with extracellular uptake and secretion rates predicted correctly for four routes in total. Thus, the results indicated the relatively higher validity of the proteomics-constrained GSMMs compared to the other data types for intracellular, compartmentalized flux predictions.
Results from GSMMs constrained with proteomic data reveal metabolic vulnerabilities of EMT
For the investigation of EMT-specific metabolic remodeling, we utilized the same methodology as before16 to identify reactions whose activity specifically requires alteration in order to switch from a epithelial flux profile (EPI) to a mesenchymal one (MES). As the proteomics-constrained GSMMs had the most accurate flux predictions, we used them for this analysis. Briefly, we used a hypergeometric test to identify whether the altered reactions were enriched with any subsystems (e.g. the metabolic pathway families with specific functional roles) within iBreast2886. The results showed that two out of the top four enriched reaction sets among EMT-linked reactions are within cholesterol metabolism (highlighted in red in Fig. 4a).
a Enrichment analysis of reactions that need alterations for the EPI to take on a MES flux phenotype in all proteomic GSMMs. This approach helps to identify reaction sets (i.e., families of pathways) which need to be altered for EMT-related changes in flux profiles. Reaction sets shown are ones with FDR-corrected p value less than 0.01. The scale (Enrichment score) represents the fraction of set reactions within the EMT reactions. The p values are FDR-adjusted p values from a hypergeometric test for enrichment of reaction sets. The two reaction sets highlighted in red are cholesterol-related. b The mechanism of inhibition of cholesterol synthesis by lovastatin. Dashed arrows represent more than one metabolic reaction. c Both D492 and D492M were treated with various concentrations of lovastatin, an inhibitor of cholesterol synthesis. The figure shows the cholesterol concentration in D492 and D492M cells after treatment with lovastatin. d Survival of D492 and D492M cells after treatment 5 µM concentration of lovastatin. Results in c) and d) are shown as mean + SEM from three experiments (shown with dots). Student’s t-test was used to estimate significance and p values were adjusted using the Benjamini–Hochberg approach.
Statins are a class of drugs that are broadly prescribed to patients with hypercholesterolemia. They work by inhibiting HMG-CoA reductase (Fig. 4b), the rate-limiting step in cholesterol synthesis26. We treated the D492 and D492M cells with lovastatin and found that following the successful inhibition of cholesterol synthesis in both cell lines (Fig. 4c), it was apparent that the D492 cells were more sensitive to the drug in terms of survival (Fig. 4d).
Thus, in addition to being the most accurate model in terms of intracellular fluxes, the analysis of the proteomic iBreast2886 GSMMs proved useful in identifying the differences in cholesterol metabolism in D492 and D492M. On the same note, we performed gene essentiality analysis of the proteomic GSMMs and found the essential genes for EPI and MES, respectively. Focusing particularly on the mesenchymal GSMM due to its metastatic involvement, we found that there were nine genes essential for the MES model. These were Argininosuccinate Lyase (ASL), Ornithine Aminotransferase (OAT), Pyruvate Dehydrogenase Complex Component X (PDHX), Proline Dehydrogenase 1 (PRODH), Renin binding protein (RENBP), Isocitrate Dehydrogenase 2 (IDH2), Guanylate Kinase 1 (GUK1), 6-Phosphogluconolactonase (PGLS) and Cystathionine Gamma-Lyase (CTH).
In order to narrow down the list of genes to verify in vitro, we evaluated the genes‘ relationship to survival of patients with claudin-low breast cancer, which is representative for the mesenchymal-like phenotype of breast cancer that expresses high levels of EMT markers27. This we achieved by measuring the concordance index (C-index) for the genes, which is a metric for predictive ability of survival models based on gene expression levels28. ASL had the highest C-index (and lowest associated p value) among the genes (Fig. 5a) and was chosen for in vitro survival analysis.
a Concordance index (C-index) of proteomic MES essential genes for overall survival prediction of patients with claudin-low breast tumors. Lines represent 95% confidence intervals and p values are from the calculation of the C-index using Noether’s method79. Genes are plotted in descending order based on the p values. b Expression levels of ASL in D492 and D492M after siRNA-mediated knockdown of the gene. Two different siRNA constructs were used for ASL (ASL-1 and ASL-2) (c) siRNA-mediated knockdown of ASL and its effects on the 96 h survival of D492 and D492M. Results in (b) and (c) are shown as mean + SEM from three experiments (shown with dots). Student’s t-test was used to estimate significance and p values were adjusted using the Benjamini–Hochberg approach.
After lowering ASL expression by 75% using small interfering RNAs (siRNA), the survival of D492M cells was significantly diminished whereas the survival of D492 cells was not altered (Fig. 5b and c). The same effect was observed when GUK1, the gene with third-lowest p value, was silenced in the cells using siRNA (Supplementary figure 3). Importantly, no effect on survival was observed after silencing the gene coding for the neighbor reaction of ASL, argininosuccinate synthase (ASS1) (Supplementary figure 3).
iBreast2886-dependent analysis of breast cancer proteome reveals subtype-specific vulnerabilities
Building on the verification of the gene essentiality predictions, we next validated the ability of iBreast2886 to identify growth vulnerabilities in breast cancer that could potentially be exploited for diagnostic or therapeutic purposes. To achieve this, we used proteomic data from breast tumors29 as constraints for the model. Again, we chose proteomic data (instead of available transcriptomic data) based on our preliminary constraint-based analysis with D492 and D492M data which showed its relatively higher accuracy in capturing intracellular flux phenotypes compared to transcriptomic data.
We hypothesized that we would identify subtype-specific metabolic vulnerabilities of breast cancer, i.e. genes specifically essential for estrogen-receptor positive (ER-positive) and ER-negative tumors. The status of the estrogen receptor has repeatedly been shown to be a significant prognostic marker, where patients with ER-negative tumors generally have shorter survival times. After creating 55 patient-specific proteomic GSMMs, we performed gene essentiality analysis on all models. Subsequently, the ER-negative and ER-positive patient GSMMs were tested for over-representation of essential genes using empirical p value calculations (see Materials and Methods).
A single essential gene was identified for ER-negative patients: Argininosuccinate Lyase (ASL) (empirical p value = 0.0419). In order to confirm these results, we acquired the metadata for the patients through the Gene Expression Omnibus (GEO) and performed survival analyses using survival in months as time and cancer-related death as event.
Univariate and multivariable Cox proportional hazard models were constructed using age and ASL separately for patients with ER-positive and ER-negative tumors. The Cox analyses (shown in Table 1) revealed that although ASL protein levels were a predictor of death in the univariate models of patients with both ER-positive and ER-negative tumors (HR = 1.16 and 1,12; p = 0.067 and 0.049, respectively), the inclusion of age in the multivariable models attenuated the effects of ASL in the ER-positive patients (HR = 1.08, p = 0.44) but not in ER-negative patients (HR = 1.12, p = 0.062). Thus, after adjusting for confounding effector age we observed that ASL was a marginally significant predictor of cancer-related death of only ER-negative patients, which confirms the results from the gene essentiality analysis using breast cancer proteomic data and iBreast2886.
There were no genes significantly enriched for ER-positive patients, likely due to the heterogeneity of this breast cancer subtype which might require further stratification based on the status of the progesterone and HER2 receptors or genes within the PAM50 panel30.
Discussion
High-throughput molecular screening can serve to focus experimental efforts on understanding the functional consequences of molecular variation. Here we have used genome-scale metabolic network modeling to reverse this classification and prioritization strategy. Rather than using high-throughput clinical data as the basis for network analysis of generic metabolic models, we used GSMMs constrained with data from cells in culture whose metabolic phenotypes resemble breast gland development to describe the metabolic landscape of breast epithelium and identify changes in metabolism associated with breast cancer.
A comparison of the different omics data from the breast epithelial cells D492 and their mesenchymal isogenic cell line D492M revealed a low correlation of the mRNA and protein levels, compatible with literature reports on the correlation of these data types22,31,32,33. There was an even lower correlation of the two different mRNA quantification methods microarray and RNA sequencing (Fig. 1) which is in accordance with the previous studies34.
The correlation of transcriptomic and proteomic data can be compartment-specific due to the different spatiotemporal nature of the molecules35. Accordingly, after constraining iBreast2886 with all the different omics data, we found that the differences in fluxes between the proteomic- and RNA-seq-constrained models were indeed highly compartment-specific (Supplementary table 1). Three compartments (cytosol, mitochondria, and the ER) had a significant correlation of ρ > 0.37, with the ER having the highest value of 0.54. A plausible explanation is that mRNA is synthesized in the nucleus, but is subsequently exported to the rough ER where protein translation takes place. Therefore, the ER displaying high correlation of reaction activity based on proteomic and transcriptomic data is unsurprising36,37.
Multiple factors influence the consistency of proteomics and transcriptomics data, not only technical ones like experimental discrepancies and different data-producing platforms, but also biological factors like gene regulation, post-translational modification, different rates of synthesis, and availability of resources35. Our findings support that these are different based on the type of cellular compartment and show that the accuracy of GSMMs flux predictions from extracellular uptake and secretion measurements is dependent upon the transcriptomic and proteomic profiles of the cellular compartment of interest.
The compartment-dependent correlation results highlight that care must be taken in the interpretation of metabolic phenotypes from high-throughput data as these may fail to accurately represent the most fundamental parts of energy metabolism. Indeed, it was apparent that the predicted relative activity between EPI and MES was highly data type-dependent (Fig. 2) with the proteomic-constrained GSMMs predicting flux phenotypes most similar to measured pathway activity using a 1-13C-glutamine tracer (Fig. 3). Further analysis of the proteomic GSMMs was successful in proposing valid changes to D492 metabolism following EMT. The EMT-linked reaction list was enriched particularly with reactions taking part in cholesterol and squalene metabolism (Fig. 4a). As a confirmation of these predictions, we found that the cholesterol-inhibiting drug lovastatin had a significantly stronger effect on the survival of D492 than D492M cells (Fig. 4d). Cholesterol has previously been shown to be a promoter of EMT38 and the cholesterol-inhibiting drug statin has been shown to inhibit cancer invasion and metastasis39,40,41. Importantly, the differences in cholesterol metabolism of D492 and D492M were not captured by a general KEGG pathway enrichment analysis of the significantly different proteins in the cell lines (Supplementary figure 4), suggesting the presence of emergent properties of the iBreast2886 network that are biologically relevant and cannot be elucidated using a generic differential expression analysis.
Similarly, the gene essentiality analysis for the proteomics-constrained MES model suggested that argininosuccinate lyase (ASL) would be essential for D492M which was confirmed by in vitro siRNA knockdown experiments (Fig. 5). Upon knockdown of the gene, there was a 22.1% reduction in survival of D492M cells on average in contrast to only 7% in D492 cells. This level of survival reduction in D492M is comparable to results from validations in previous studies using similar methodology proposing metabolic targets, where 10-80% reduction in survival have been observed upon in vitro knockdown of the main metabolic target genes14,42. A manual investigation of the GSMM flux profiles revealed three likely reasons for the essentiality of ASL: 1) compromised proline synthesis via OAT accompanied by 2) decreased fumarate production for the TCA cycle and 3) decreased OAA to aspartate conversion that compromises aminotransferase activity and therefore anaplerotic fueling of the TCA cycle. In addition to ASL, we identified six other significant targets from the gene essentiality analysis, most of which have been associated with poor cancer survival6,43,44,45. For example, increased expression of the IDH2 gene has been shown to be overexpressed in endometrial, prostate, testicular, and advanced colon cancer46,47,48, and we have recently demonstrated that IDH2 indeed fuels reductive glutaminolysis and fatty acid synthesis in the D492 and D492M cells in an accompanying study49.
Several studies have carried out metabolic network analysis on a compendium of clinical transcriptomic or proteomic data to extract and prioritize metabolic features of importance14,15,50,51,52. Using our breast metabolic GSMM iBreast2886, we analysed publically available breast tumor proteomic data from 55 breast cancer patients and identified ASL as a metabolic vulnerability of the aggressive ER-negative breast cancer.
Thus, we found that ASL was both essential for D492M cell growth and related to the worse prognosis of ER-negative breast cancer patients. The enzyme coded by ASL, argininosuccinate lyase, produces fumarate and arginine from the breakdown of argininosuccinate. Arginine is a non-essential proteogenic amino acid involved in nitrogen detoxification and the generation of nitric oxide (NO) which is important for invasion and metastasis in various cancer types6,53. Downregulation of ASL has been shown to inhibit the growth of breast cancer tumors in vitro and in vivo54. Rabinovich et al.55 reported that downregulation of ASS1, an enzyme directly upstream of ASL, increased pyrimidine synthesis and cancer cell proliferation but did not see the same connection with ASL. The different effect of siRNA knockdown of ASL and ASS1 on D492 and D492M survival reported here support a mutually exclusive relationship of ASL and ASS1 as only ASL and not ASS1 was found to be essential for growth of D492M (Fig. 5c and Supplementary figure 3). This however does not explain the observed differences in the context of linear pathway flux within iBreast2886. A possible explanation is a secondary function of ASL, as it has been shown to influence cyclin A2 levels by direct binding in hepatocellular carcinoma, independent of its enzymatic activity within the ASS1-ASL node that also promoted anchorage-independent growth56. Intercellular exchange of argininosuccinate between ASL- and ASS1-deficient cells, as demonstrated by Davidson et al.57, furthermore indicates that the two enzymes need not be co-regulated within a single-cell type. This type of tissue-level metabolic crosstalk would not be captured by our single-cell metabolic reconstruction iBreast2886. Nevertheless, the components of the ASS1-ASL node, citrulline, and fumarate, have been reported to be significantly lower and higher, respectively, in ER-negative breast cancer compared to ER-positive which supports altered activity within the ASS1-ASL metabolic node58. The findings additionally support more studies that have shown that metabolic vulnerabilities of breast cancer lie within arginine metabolism54,59,60.
Taken together, the study demonstrates that the metabolism of EMT captured within iBreast2886 is practical for data integration and analysis and that proposed phenotypes are in agreement both with prior investigations of EMT/metastasis and ER-negative breast cancer metabolism. The iBreast2886 reconstruction is first and foremost a metabolic model descriptive of the steady-state metabolic phenotypes that the D492 EMT cell model can achieve based upon the integration of mRNA transcription, protein translation, and metabolite uptake and secretion rates. The integrated analysis of multiple iBreast2886 GSMMs constrained with separate data types collectively yielded more accurate predictions than each individual GSMM, as shown here with the EMT-related changes in cholesterol metabolism and ASL essentiality.
Limitations of iBreast2886 include lack of actual measurements of fatty acid oxidation and cholesterol uptake/secretion rates which might further increase predictive accuracy of iBreast2886. Genes involved in the oxidation of fatty acids are known to correlate with reduced cancer patient survival14,61,62 and the relationship of cholesterol to EMT and metastasis has been discussed here above38,39,40,41. The robustness and plasticity of breast tissue metabolism are also more complex than is captured by iBreast2886, which is solely based upon one EMT cell culture model and media constraints that may not accurately reflect the breast tissue microenvironment63 and lack flux extremities that may arise from kinetic regulation. Steps towards further understanding of EMT metabolism could be performed by expanding iBreast2886 to account for additional cell lines alongside focused studies aimed at addressing metabolic gaps and network inconsistencies whose presence was demonstrated in this study using isotope tracer analysis. In this way, biochemically accurate descriptions of EMT metabolism in breast tissue to aid in translational cancer research may be pushed forward.
Materials and methods
Cell culture
D492 and D492M were cultured in a serum-free H14 medium at 37°C and 5% CO2 as previously described19. H14 is a fully defined medium comprised of DMEM/F12 base with 250 ng/ml insulin, 10 µg/ml transferrin, 10 ng/ml EGF, 2.6 ng/ml sodium selenite, 10-10 M estradiol, 1.4 µM hydrocortisone, 7.1 ng/ml prolactin, 100 IU penicillin, 0.1 mg/ml streptomycin and 2 mM glutamine. Medium was changed every 48 h while propagating cells, and experiments were performed within four passages. D492 and D492M cells were kindly provided by the Stem Cell Research Unit, University of Iceland, and were screened for Mycoplasma infections monthly using PCR-based tests at the Biomedical Center, University of Iceland.
Origin of iBreast2886 GSMM for breast metabolism
Genome-scale metabolic model construction and analysis were carried out in MATLAB using the COBRA Toolbox64. The genome-scale breast tissue metabolic model from Halldorsson et al.16 was used as a base model. Briefly, RNA sequencing data from both the D492 and D492M cell lines19 was used to create a metabolic model specific for breast tissue. To achieve this, the human metabolic reconstruction Recon 2 was employed65. All genes in the RNA sequencing data with expression values exceeding a fixed cut-off value (1 RPM) in either cell line were identified. To identify the metabolic reactions associated with the list of genes, the Gene-Protein Rules (GPRs) of Recon 2 were used. The FASTCORE model building algorithm66 was subsequently applied to build a functional metabolic network from the list of identified reactions. The resulting network, referred to as the iBreast2886 model, was manually curated to ensure no metabolites or pathways were blocked or missing.
Construction and analysis of cell-type-specific epithelial and mesenchymal GSMMs
The iBreast2886 reconstruction was used to create cell-type-specific models of epithelial D492 and mesenchymal D492M based on microarray, proteomic, and RNA sequencing (RNA-seq) data. The workflow of the model construction is outlined in Supplementary figure 5. Briefly, the genes/proteins from each dataset (along with cell-type-specific uptake and secretion rates of multiple metabolites in the media) were used to constrain iBreast2886 to create two models (EPI for epithelial D492, and MES for mesenchymal D492M). Furthermore, the fourth pair of EPI and MES was added that did not contain any intracellular constraints imposed by omics data, but only the cell-type-specific uptake and secretion rates. This gave rise to four pairs of EPI and MES models, where each EPI model shared the same stoichiometry and uptake/secretion rates but had different intracellular reaction constraints based on the different datasets. The same applied to the MES models. See Supplementary information for details.
Stable isotope tracing analysis
D492 and D492M cells were cultured until confluent as described above. The medium was then changed to a complete H14 containing 1-13C-labeled glutamine (Cambridge Isotope Laboratories, Inc., MA, USA). After 6 h of culturing with the 13C-labeled carbon source, cell metabolism was quenched using cold methanol and intracellular metabolites were extracted using ACN extraction67. Analyses were performed on a UPLC system as described in Rolfsson et al.68. Results were presented as the total contribution (TC) of carbon sources to measured metabolites69:
Where n is the number of C atoms in the metabolite, i represents the isotopologues and m is the relative fraction of the isotopologues.
Comparison of GSMM flux predictions and 13C-labeling profiles
Individual flux distributions from constraint-based modeling of GSMMs were used to estimate the relative contribution of extracellular metabolites to intracellular metabolites of interest. This approach is suitable to measure the flow of carbons between metabolites within GSMMs to ultimately quantify the total activity of specific metabolic routes within pathways that can subsequently be directly compared to definitive results from stable isotope tracer analyses. A schematic explaining the metabolic route activity measure is shown in Supplementary figure 6. A single flux distribution represent the flux values of all reactions within a GSMM which is subject to the constraints applied to the model. In order to calculate the relative flux value \(v_{rel}\) from metabolite \(m_i\) to \(m_{i + 1}\) within a pathway of interest, we first identify all consuming reactions of metabolite \(m_i\) using the stoichiometric matrix S. Then, for a single flux distribution, one can calculate the sum of consuming flux of metabolite \(m_i\), and the relative flux that is used to produce only metabolite \(m_{i + 1}\), which we will call \(v_{rel}\). If k is a consuming reaction of a particular metabolite of interest, then the \(v_{rel}\) value for k is calculated from the raw flux value of k divided by the sum of the fluxes of all K reactions consuming the same metabolite as k. Therefore, the \(v_{rel}\) of k (or \(v_{rel}(k)\)) in a single flux vector is calculated as follows:
Where \(w_{comp}\) is the weight given to the relative flux value based on the relative abundance of the compartment it takes place in, since some reactions take place in more than one compartment. The \(v_{rel}(k)\) values for all transport reactions were assumed to be 1. The weights for the compartments were as follows: Cytosol 0.54, mitochondria 0.22, ER 0.12, nucleus 0.06, golgi apparatus 0.03, peroxisomes and lysosomes 0.01, and are representative of their relative volume within cells in general70.
Using the relative consumption values for a list of reactions within a single flux vector, it is possible to calculate the metabolic route activity (MRA). To calculate the MRA from metabolite m to m + k, calculate the sum of the log of relative flux values (from Eq. (2)) within that route:
Where the first reaction is the consumption of metabolite \(m_i\) to produce metabolite \(m_{i + 1}\). The MRA of multiple flux vectors (e.g., within a random sampling matrix) can be calculated to get a distribution of MRA within a specific constrained GSMM.
Lovastatin assay
D492 and D492M cells were treated with 5, 10, and 100 µM concentration of lovastatin (Tocris Bioscience, Bristol, UK) for 24 h after which both cholesterol abundance and cell numbers were assessed. The cholesterol was measured using Amplex™ Red Cholesterol Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA) according to manufacturers protocol. The cell numbers were assessed using crystal violet staining. Briefly, after 24 h of treatment, the cells were fixed using ice-cold methanol and stained with crystal violet (0.5%). The stain was subsequently released using 10% acetic acid and absorption was measured at 570 nm.
Scoring of in silico gene essentiality candidates
The METABRIC breast cancer clinical dataset71 was downloaded from cBioPortal72,73. The clinical metadata includes information about the claudin-status of the tumors in the data. Using only patients with tumors classified as claudin-low and available survival data (n = 199), we performed a survival analysis. In short, patients were split into two groups based on the best-splitting expression level (as identified through R-package survminer’s surv_cutpoint() function) of a gene of interest. The prognoses of the groups were then examined by calculating the concordance index (C-index)28, which provides an overall measure of predictive accuracy of the genes’ expression level with right-censored survival data.
Small interfering RNA (siRNA) knockdown experiments
For the knockdown experiments, Silencer Select siRNAs (Thermo) were used (Negative Control No 1 #4390843), ASL (s1669 and s1671), and ASS1 (s1684). Cells were seeded at density of 3000 cells/well in a 96 well plate. Prior to seeding, the 96 well plates were coated with siRNA and Lipofectamine RNAiMAX (Thermo) for 15 min. Final concentration of siRNA in each well, after addition of cells, was 10 nM. Transfected cells were incubated at 37 °C and 5% CO2 for 96 h at the end of which cell survival and gene expression were assessed. To measure cell survival, CellTiter Glo Luminescent Cell Viability Assay (Promega, Madison, WI, USA) was used according to instructions of the manufacturer. SpectraMax plate reader was used to measure luminescence at 560 nm. To measure the gene expression, real time quantitative polymerase chain reaction (qPCR) was used.
Real-time PCR
Total RNA was isolated using TRI-Reagent (Thermo) according to the manufacturer’s instructions. RNA concentration was measured using NanoDrop One (Thermo). 0.4 to 1 ug RNA was reverse transcribed to cDNA using High-Capacity cDNA Reverse Transcription kit (Thermo). Real-time quantitative PCR reactions were carried out using Luna Universal qPCR Master Mix (New England Biolabs, Ipswich, MA, USA) according to manufacturer’s instructions on a BioRad CFX384 Touch™ Real Time System (BioRad Laboratories, Hercules, CA, USA). Gene expression levels were determined with CFX Manager Software (BioRad). Primers were designed using the Primer3 software74. Primers spanning exon junctions were chosen to ensure specificity. Differences in relative expression were estimated using the 2∆∆Ct method. The primer sequences used for quantifying the gene expression were: ASL-fwd 5‘-GGAAGCTGTGTTTGAAGTGTCA-3‘, ASL-rev 5‘-CCATGTTCTCTTGGTGAATCTG-3‘, ASS1-fwd 5‘-CAGGAAAGGGGAACGATCAGGT-3‘, ASS1-rev 5‘-CGTGTTGCTTTGCGTACTCCAT-3‘, GUK1-fwd 5‘-CTTCATCGAGCATGCCGAGTTC-3‘, GUK1-rev 5‘-GAACCTGTATGGCACGAGCAAG-3‘, ACTB-fwd 5‘-CTTCCTGGGTGAGTGGAGACTG-3‘ and ACTB-rev 5‘-GAGGGAAATGAGGGCAGGACTT-3‘.
Analysis of clinical breast cancer data using iBreast2886
Proteomic breast cancer data were acquired from Tang et al.29. After removing identifers with missing data in more than 20% of samples, the data were imported into MATLAB for constraint-based modeling.
Patient-specific GSMMs were constructed from iBreast2886, where the reactions were only constrained in a patient model if their associated protein levels were below the 60th percentile in all patients. The same amount of constraint was applied to the selected reactions as for the EPI and MES models (as described above). The median percentage of constrained reactions in the patients was 3.8%. Gene essentiality analysis was carried out using FBA as described above.
Essential genes that were over-represented in the GSMMs of estrogen receptor (ER) negative (n1 = 33) and positive patients (n2 = 32) were identified by randomly sampling n1 and n2 patient-specific GSMMs 1000 times from the whole GSMM list. Then, an empirical p value (\(\hat p\)) was calculated for each gene in the ER-negative and ER-positive patient subsets using the formula from North et al.75:
Where \(\hat p\) is the empirical p value, n is the number of resampled sets (1000 in this case) and r is the number of times the resampled sets have an equal or greater number of an essential gene compared to the ER-negative or ER-positive patient sets.
Genes with a \(\hat p\) < 0.05 were identified and their proteomic levels29 were tested as subtype-specific survival predictors using the patient metadata acquired from GEO (GSE37751). The metadata used were cancer-related death and survival in months that were acquired using the R-package GEOquery76. To assess the effects of genes and confounding variables on patient survival, Cox-proportional hazard models were employed using the R-package survival77.
Statistical analysis
For comparison of two groups, a two-sided Student’s t-test was employed unless when the data did not follow a normal distribution, when the non-parametric Mann–Whitney U-test was used. When more than a single treatment was compared in the cell lines, the treatments were all compared to the negative control using two-sided Student’s t-test and subsequently, the p values were adjusted for multiple comparisons using the Benjamini–Hochberg approach. For the comparison of two distributions (e.g., in the metabolic route activity measurements), a Kolmogorov–Smirnov test was used. Presented data were from at least three independent experiments (represented by dots) and were summarized using mean + standard error. The asterisks in each figure represent the p values (*<0.05, **<0.01, ***<0.001, ****<0.0001, ns = not significant). Statistical methods used for GSMM analysis of breast cancer patients are described in the Analysis of clinical breast cancer data using iBreast2886 section. All statistical analysis was carried out using the R programming language78.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request. The breast cancer proteomic data that were analysed during the current study are available in Tang et al.29 and the metadata were acquired from the Gene Expression Omnibus (GEO), accession no. GSE37751.
Code availability
All code used in this study for constraint-based modeling and analysis are accessible at https://github.com/siggitrausti/EMT_metabolic_modeling.
References
Chaffer, C. L. & Weinberg, R. A. A perspective on cancer cell metastasis. Science 331, 1559–1564 (2011).
Tseng, P.-L. et al. The decrease of glycolytic enzyme hexokinase 1 accelerates tumor malignancy via deregulating energy metabolism but sensitizes cancer cells to 2-deoxyglucose inhibition. Oncotarget 9, 18949–18969 (2018).
Wei, L. et al. Leptin promotes epithelial-mesenchymal transition of breast cancer via the upregulation of pyruvate kinase M2. J. Exp. Clin. Cancer Res. 35, 166 (2016).
Liu, W.-S. et al. Isocitrate dehydrogenase 1-snail axis dysfunction significantly correlates with breast cancer prognosis and regulates cell invasion ability. Breast Cancer Res. 20, 25 (2018).
Liu, H., Huang, X. & Ye, T. MiR-22 down-regulates the proto-oncogene ATP citrate lyase to inhibit the growth and metastasis of breast cancer. Am. J. Transl. Res. 10, 659–669 (2018).
Elia, I. et al. Proline metabolism supports metastasis formation and could be inhibited to selectively target metastasizing cancer cells. Nat. Commun. 8, 15267 (2017).
Elia, I., Doglioni, G. & Fendt, S. M. Metabolic hallmarks of metastasis formation. Trends Cell Biol. 28, 673–684 (2018).
Kalluri, R. & Weinberg, R. A. The basics of epithelial-mesenchymal transition. J. Clin. Invest 119, 1420–1428 (2009).
Ye, X. & Weinberg, R. A. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 25, 675–686 (2015).
Liao, T. T. & Yang, M. H. Revisiting epithelial-mesenchymal transition in cancer metastasis: the connection between epithelial plasticity and stemness. Mol. Oncol. 11, 792–804 (2017).
Hanahan, D. & Weinberg, R. A. Hallmarks of cancer: the next generation. Cell 144, 646–674 (2011).
Mardinoglu, A. & Nielsen, J. Editorial: the impact of systems medicine on human health and disease. Front Physiol. 7, 552 (2016).
Yizhak, K., Gabay, O., Cohen, H. & Ruppin, E. Model-based identification of drug targets that revert disrupted metabolism and its application to ageing. Nat. Commun. 4, 2632 (2013).
Yizhak, K. et al. Phenotype-based cell-specific metabolic modeling reveals metabolic liabilities of cancer. eLife 3, e03641 (2014).
Jerby, L. et al. Metabolic associations of reduced proliferation and oxidative stress in advanced breast cancer. Cancer Res. 72, 5712–5720 (2012).
Halldorsson, S. et al. Metabolic re-wiring of isogenic breast epithelial cell lines following epithelial to mesenchymal transition. Cancer Lett. 396, 117–129 (2017).
Tam, W. L. et al. Protein kinase C α is a central signaling node and therapeutic target for breast cancer stem cells. Cancer Cell 24, 347–364 (2013).
Ackland, M. L. et al. Epidermal growth factor-induced epithelio-mesenchymal transition in human breast carcinoma. Cells Lab. Investig. 83, 435–448 (2003).
Sigurdsson, V. et al. Endothelial induced EMT in breast epithelial cells with stem cell properties. PLoS ONE 6, e23833 (2011).
Shaul, Y. D. et al. Dihydropyrimidine accumulation is required for the epithelial-mesenchymal transition. Cell 158, 1094–1109 (2014).
Jiang, L. et al. Reductive carboxylation supports redox homeostasis during anchorage-independent growth. Nature 532, 255–258 (2016).
Maier, T., Güell, M. & Serrano, L. Correlation of mRNA and protein in complex biological samples. FEBS Lett. 583, 3966–3973 (2009).
Patil, K. R. & Nielsen, J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc. Natl Acad. Sci. USA 102, 2685 (2005).
Bordbar, A., Monk, J. M., King, Z. A. & Palsson, B. O. Constraint-based models predict metabolic and associated cellular functions. Nat. Rev. Genet. 15, 107–120 (2014).
Schellenberger, J. & Palsson, B. O. Use of randomized sampling for analysis of metabolic networks. J. Biol. Chem. 284, 5457–5461 (2009).
Istvan, E. S. & Deisenhofer, J. Structural mechanism for statin inhibition of HMG-CoA reductase. Science 292, 1160–1164 (2001).
Prat, A. et al. Phenotypic and molecular characterization of the claudin-low intrinsic subtype of breast cancer. Breast Cancer Res. 12, R68–R68 (2010).
Harrell, F. E., Califf, R. M., Pryor, D. B., Lee, K. L. & Rosati, R. A. Evaluating the yield of medical tests. JAMA J. Am. Med. Assoc. 247, 2543–2546 (1982).
Tang, W. et al. Integrated proteotranscriptomics of breast cancer reveals globally increased protein-mRNA concordance associated with subtypes and survival. Genome Med. 10, 94 (2018).
Parker, J. S. et al. Supervised risk predictor of breast cancer based on intrinsic subtypes. J. Clin. Oncol. 27, 1160–1167 (2009).
Edfors, F. et al. Gene-specific correlation of RNA and protein levels in human cells and tissues. Mol. Syst. Biol. 12, 883 (2016).
Darmanis, S. et al. Simultaneous multiplexed measurement of RNA and proteins in single cells. Cell Rep. 14, 380–389 (2016).
Gong, H. et al. Single-cell protein-mRNA correlation analysis enabled by multiplexed dual-analyte co-detection. Sci. Rep. 7, 2776 (2017).
Trost, B. et al. Concordance between RNA-sequencing data and DNA microarray data in transcriptome analysis of proliferative and quiescent fibroblasts. R. Soc. Open Sci. 2, 150402 (2015).
Liu, Y., Beyer, A. & Aebersold, R. On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016).
Lu, R. et al. Systems-level dynamic analyses of fate change in murine embryonic stem cells. Nature 462, 358–362 (2009).
Cooper, G. M. The Cell - A Molecular Approach 2nd Edition. (Sunderland (MA): Sinauer Associates, 2000). citeulike-article-id:10266975.
Alikhani, N. et al. Mammary tumor growth and pulmonary metastasis are enhanced in a hyperlipidemic mouse model. Oncogene 32, 961–967 (2013).
Taras, D. et al. Pravastatin reduces lung metastasis of rat hepatocellular carcinoma via a coordinated decrease of MMP expression and activity. J. Hepatol. 46, 69–76 (2007).
Nielsen, S. F., Nordestgaard, B. G. & Bojesen, S. E. Statin use and reduced cancer-related mortality. N. Engl. J. Med. 367, 1792–1802 (2012).
Zhang, J. et al. Statins, autophagy and cancer metastasis. Int. J. Biochem. Cell Biol. 45, 745–752 (2013).
Dai, Z. et al. Identification of cancer-associated metabolic vulnerabilities by modeling multi-objective optimality in metabolism. Cell Commun. Signal. 17, 124 (2019).
Chen, B. et al. MicroRNA-26a regulates glucose metabolism by direct targeting PDHX in colorectal cancer cells. BMC Cancer 14, 443 (2014).
da Rocha, A. A. et al. Hepatocyte growth factor-regulated tyrosine kinase substrate (HGS) and guanylate kinase 1 (GUK1) are differentially expressed in GH-secreting adenomas. Pituitary 9, 83–92 (2006).
Liu, Y. et al. Ornithine aminotransferase promoted the proliferation and metastasis of non-small cell lung cancer via upregulation of miR-21. J. Cell. Physiol. 234, 12828–12838 (2019).
Guirguis, A. et al. Use of gene expression profiles to stage concurrent endometrioid tumors of the endometrium and ovary. Gynecol. Oncol. 108, 370–376 (2008).
Altenberg, B. & Greulich, K. O. Genes of glycolysis are ubiquitously overexpressed in 24 cancer classes. Genomics 84, 1014–1020 (2004).
Lv, Q. et al. Altered expression levels of IDH2 are involved in the development of colon cancer. Exp. Ther. Med. 4, 801–806 (2012).
Trausti Karvelsson, S. et al. Altered glutamine metabolism exposes EMT derived mesenchymal cells to PI3K/Akt/mTOR pathway inhibition. (2020) https://doi.org/10.21203/RS.3.RS-100299/V1.
Bjornson, E. et al. Stratification of hepatocellular carcinoma patients based on acetate utilization. Cell Rep. 13, 2014–2026 (2015).
Benfeitas, R. et al. Characterization of heterogeneous redox responses in hepatocellular carcinoma patients using network analysis. EBioMedicine (2018) https://doi.org/10.1016/j.ebiom.2018.12.057.
Mardinoglu, A. et al. Integration of clinical data with a genome-scale metabolic model of the human adipocyte. Mol. Syst. Biol. 9, (2013).
Chen, L.-H., Hsu, W.-L., Tseng, Y.-J., Liu, D.-W. & Weng, C.-F. Involvement of DNMT 3B promotes epithelial-mesenchymal transition and gene expression profile of invasive head and neck squamous cell carcinomas cell lines. BMC Cancer 16, 431 (2016).
Huang, H. L. et al. Argininosuccinate lyase is a potential therapeutic target in breast cancer. Oncol. Rep. 34, 3131–3139 (2015).
Rabinovich, S. et al. Diversion of aspartate in ASS1-deficient tumours fosters de novo pyrimidine synthesis. Nature 527, 379–383 (2015).
Hung, Y.-H. et al. Argininosuccinate lyase interacts with cyclin A2 in cytoplasm and modulates growth of liver tumor cells. Oncol. Rep. 37, 969–978 (2017).
Davidson, J. S., Baumgarten, I. M. & Harley, E. H. Metabolic cooperation between argininosuccinate synthetase and argininosuccinate lyase deficient human fibroblasts. Exp. Cell Res 150, 367–378 (1984).
Budczies, J. et al. Comparative metabolomics of estrogen receptor positive and estrogen receptor negative breast cancer: alterations in glutamine and beta-alanine metabolism. J. Proteom. 94, 279–288 (2013).
Qiu, F. et al. Arginine starvation impairs mitochondrial respiratory function in ASS1-deficient breast cancer cells. Sci. Signal. 7, ra31–ra31 (2014).
Cheng, C.-T. et al. Arginine starvation kills tumor cells through aspartate exhaustion and mitochondrial dysfunction. Commun. Biol. 1, 1–15 (2018).
Jia, M. et al. Estrogen receptor α promotes breast cancer by reprogramming choline metabolism. Cancer Res. 76, 5634–5646 (2016).
Aiderus, A., Black, M. A. & Dunbier, A. K. Fatty acid oxidation is associated with proliferation and prognosis in breast and other cancers. BMC Cancer 18, 805 (2018).
Katzir, R. et al. The landscape of tiered regulation of breast cancer cell metabolism. Sci. Rep. 9, 1–12 (2019).
Heirendt, L. et al. Creation and analysis of biochemical constraint-based models using the COBRA Toolbox v.3.0. Nat. Protoc. 14, 639–702 (2019).
Thiele, I. et al. A community-driven global reconstruction of human metabolism. Nat. Biotechnol. 31, 419–425 (2013).
Vlassis, N., Pacheco, M. P. & Sauter, T. Fast reconstruction of compact context-specific metabolic network models. PLOS Comput. Biol. 10, e1003424 (2014).
Paglia, G. et al. Intracellular metabolite profiling of platelets: evaluation of extraction processes and chromatographic strategies. J. Chromatogr. B 898, 111–120 (2012).
Rolfsson, Ó. et al. Mannose and fructose metabolism in red blood cells during cold storage in SAGM. Transfusion 57, 2665–2676 (2017).
Buescher, J. M. et al. A roadmap for interpreting (13)C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 34, 189–201 (2015).
Alberts, B. et al. Essential cell biology. (Garland Science, 2013).
Pereira, B. et al. The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 (2016).
Cerami, E. et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Disco. 2, 401–404 (2012).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1 (2013).
Untergasser, A. et al. Primer3–new capabilities and interfaces. Nucleic Acids Res. 40, e115–e115 (2012).
North, B. V., Curtis, D. & Sham, P. C. A note on the calculation of empirical P values from Monte Carlo procedures [1]. Am. J. Hum. Genet. 71, 439–441 (2002).
Sean, D. & Meltzer, P. S. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 23, 1846–1847 (2007).
Therneau, T. M. & Grambsch, P. M. Modeling Survival Data: Extending the Cox Model. (Springer New York, 2000). https://doi.org/10.1007/978-1-4757-3294-8.
R Core Team. R: A Language and Environment for Statistical Computing. (2020).
Penciana, M. J. & D’Agostino, R. B. Overall C as a measure of discrimination in survival analysis: model specific population value and confidence interval estimation. Stat. Med. 23, 2109–2123 (2004).
Acknowledgements
This work was supported by the Icelandic Research Fund (#163254-051), Göngum Saman, and the Norwegian Research Council (#239940). The authors thank Freyr Johannsson and Sarah McGarrity for valuable input regarding constraint-based modeling methodology, 13C isotope tracer, and metabolomics analysis.
Author information
Authors and Affiliations
Contributions
Conceptualization, S.T.K., S.G. and O.R.; Methodology, S.T.K., A.R., Q.W., S.A.M., S.H., S.G. and O.R.; Software, S.T.K. and S.G.; Formal analysis and Investigation, S.T.K., A.S., Q.W., S.A.M.; Data curation, S.T.K., Q.W., A.S. and S.H.; Visualization, S.T.K.; Writing – Original Draft, S.T.K. and O.R.; Writing––Review and Editing, all authors; Funding acquisition, G.M.M., S.A.M., S.G. and O.R., Supervision, G.M.M., S.A.M., S.G. and O.R.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Karvelsson, S.T., Wang, Q., Hilmarsdottir, B. et al. Argininosuccinate lyase is a metabolic vulnerability in breast development and cancer. npj Syst Biol Appl 7, 36 (2021). https://doi.org/10.1038/s41540-021-00195-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41540-021-00195-5