Abstract
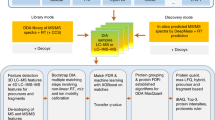
Here we describe the use of data-independent acquisition (DIA) on a Q-Exactive mass spectrometer for the detection and quantification of peptides in complex mixtures using the Skyline Targeted Proteomics Environment (freely available online at http://skyline.maccosslab.org). The systematic acquisition of mass spectrometry (MS) or tandem MS (MS/MS) spectra by DIA is in contrast to DDA, in which the acquired MS/MS spectra are only suitable for the identification of a stochastically sampled set of peptides. Similarly to selected reaction monitoring (SRM), peptides can be quantified from DIA data using targeted chromatogram extraction. Unlike SRM, data acquisition is not constrained to a predetermined set of target peptides. In this protocol, a spectral library is generated using data-dependent acquisition (DDA), and chromatograms are extracted from the DIA data for all peptides in the library. As in SRM, quantification using DIA data is based on the area under the curve of extracted MS/MS chromatograms. In addition, a quality control (QC) method suitable for DIA based on targeted MS/MS acquisition is detailed. Not including time spent acquiring data, and time for database searching, the procedure takes ∼1–2 h to complete. Typically, data acquisition requires roughly 1–4 h per sample, and a database search will take 0.5–2 h to complete.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout















Similar content being viewed by others
References
Purvine, S., Eppel, J.-T., Yi, E.C. & Goodlett, D.R. Shotgun collision-induced dissociation of peptides using a time-of-flight mass analyzer. Proteomics 3, 847–850 (2003).
Venable, J.D., Dong, M.-Q., Wohlschlegel, J., Dillin, A. & Yates, J.R. Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods 1, 39–45 (2004).
Law, K.P. & Lim, Y.P. Recent advances in mass spectrometry: data independent analysis and hyper reaction monitoring. Expert Rev. Proteomics 10, 551–566 (2013).
Gillet, L.C. et al. Targeted data extraction of the MS/MS spectra generated by data-independent acquisition: a new concept for consistent and accurate proteome analysis. Mol. Cell. Proteomics 11, O111.016717 (2012).
Stahl-Zeng, J. et al. High sensitivity detection of plasma proteins by multiple reaction monitoring of N-glycosites. Mol. Cell. Proteomics 6, 1809–1817 (2007).
Lange, V., Picotti, P., Domon, B. & Aebersold, R. Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 222 (2008).
Percy, A.J., Chambers, A.G., Yang, J., Hardie, D.B. & Borchers, C.H. Advances in multiplexed MRM-based protein biomarker quantitation toward clinical utility. Biochim. Biophys. Acta 1844, 917–926 (2014).
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Chait, B.T. Mass spectrometry: bottom-up or top-down? Science 314, 65–66 (2006).
Yates, J.R., Ruse, C.I. & Nakorchevsky, A. Proteomics by mass spectrometry: approaches, advances, and applications. Annu. Rev. Biomed. Eng. 11, 49–79 (2009).
Payne, A.H. & Glish, G.L. in Methods in Enzymology (ed. Burlingame, A.L.) 402, 109–148 (Academic Press, 2005).
McLafferty, F.W. Tandem Mass Spectrometry (John Wiley & Sons Inc., 1983).
Egertson, J.D. et al. Multiplexed MS/MS for improved data-independent acquisition. Nat. Methods 10, 744–746 (2013).
Norbeck, A.D., Monroe, M.E., Adkins, J.N. & Smith, R.D. The utility of accurate mass and LC elution time information in the analysis of complex proteomes. J. Am. Soc. Mass Spectrom. 16, 1239–1249 (2005).
Weisbrod, C.R., Eng, J.K., Hoopmann, M.R., Baker, T. & Bruce, J.E. Accurate peptide fragment mass analysis: multiplexed peptide identification and quantification. J. Proteome Res. 11, 1621–1632 (2012).
Clough, T., Thaminy, S., Ragg, S., Aebersold, R. & Vitek, O. Statistical protein quantification and significance analysis in label-free LC-MS experiments with complex designs. BMC Bioinformatics 13, S6 (2012).
MacLean, B. et al. Skyline: an open-source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics 26, 966–968 (2010).
Wang, J., Bourne, P.E. & Bandeira, N. MixGF: spectral probabilities for mixture spectra from more than one peptide. Mol. Cell Proteomics 13, 3688–3697 (2014).
Röst, H.L. et al. OpenSWATH enables automated, targeted analysis of data-independent acquisition MS data. Nat. Biotechnol. 32, 219–223 (2014).
Gallien, S., Duriez, E., Demeure, K. & Domon, B. Selectivity of LC-MS/MS analysis: Implication for proteomics experiments. J. Proteomics 81, 148–158 (2013).
Matthews, D.E. & Hayes, J.M. Systematic errors in gas chromatography-mass spectrometry isotope ratio measurements. Anal. Chem. 48, 1375–1382 (1976).
King, N.L. et al. Analysis of the Saccharomyces cerevisiae proteome with PeptideAtlas. Genome Biol. 7, R106 (2006).
Canterbury, J.D., Merrihew, G.E., MacCoss, M.J., Goodlett, D.R. & Shaffer, S.A. Comparison of data acquisition strategies on quadrupole ion trap instrumentation for shotgun proteomics. J. Am. Soc. Mass Spectrom. 25, 2048–2059 (2014).
Scigelova, M., Hornshaw, M., Giannakopulos, A. & Makarov, A. Fourier transform mass spectrometry. Mol. Cell Proteomics 10, M111.009431 (2011).
Jonscher, K.R. & Yates, J.R. III. The quadrupole ion trap mass spectrometer—a small solution to a big challenge. Anal. Biochem. 244, 1–15 (1997).
Clauser, K.R., Baker, P. & Burlingame, A.L. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal. Chem. 71, 2871–2882 (1999).
Zubarev, R.A., Håkansson, P. & Sundqvist, B. Accuracy requirements for peptide characterization by monoisotopic molecular mass measurements. Anal. Chem. 68, 4060–4063 (1996).
Marshall, A.G., Hendrickson, C.L. & Jackson, G.S. Fourier transform ion cyclotron resonance mass spectrometry: a primer. Mass Spectrom. Rev. 17, 1–35 (1998).
Randall, S.M., Cardasis, H.L. & Muddiman, D.C. Factorial experimental designs elucidate significant variables affecting data acquisition on a quadrupole orbitrap mass spectrometer. J. Am. Soc. Mass Spectrom. 24, 1501–1512 (2013).
Eng, J.K., McCormack, A.L. & Yates, J.R. III. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Käll, L., Canterbury, J.D., Weston, J., Noble, W.S. & MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 4, 923–925 (2007).
Perry, R.H., Cooks, R.G. & Noll, R.J. Orbitrap mass spectrometry: instrumentation, ion motion and applications. Mass Spectrom. Rev. 27, 661–699 (2008).
Plumb, R.S. et al. UPLC/MS(E); a new approach for generating molecular fragment information for biomarker structure elucidation. Rapid Commun. Mass Spectrom. 20, 1989–1994 (2006).
Moon, M.H., Myung, S., Plasencia, M., Hilderbrand, A.E. & Clemmer, D.E. Nanoflow LC/ion mobility/CID/TOF for proteomics: analysis of a human urinary proteome. J. Proteome Res. 2, 589–597 (2003).
Yost, R.A. & Enke, C.G. Selected ion fragmentation with a tandem quadrupole mass spectrometer. J. Am. Chem. Soc. 100, 2274–2275 (1978).
Yost, R.A. & Enke, C.G. Triple-quadrupole mass spectrometry for direct mixture analysis and structure elucidation. Anal. Chem. 51, 1251–1264 (1979).
Bensimon, A., Heck, A.J.R. & Aebersold, R. Mass spectrometry–based proteomics and network biology. Annu. Rev. Biochem. 81, 379–405 (2012).
Stahl, D.C., Swiderek, K.M., Davis, M.T. & Lee, T.D. Data-controlled automation of liquid chromatography/tandem mass spectrometry analysis of peptide mixtures. J. Am. Soc. Mass Spectrom. 7, 532–540 (1996).
Bantscheff, M., Schirle, M., Sweetman, G., Rick, J. & Kuster, B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 (2007).
McLafferty, F.W. Tandem mass spectrometry. Science 214, 280–287 (1981).
Anderson, N.L. & Anderson, N.G. The human plasma proteome history, character, and diagnostic prospects. Mol. Cell Proteomics 1, 845–867 (2002).
Geromanos, S.J. et al. The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data-dependent LC-MS/MS. Proteomics 9, 1683–1695 (2009).
Bern, M. et al. Deconvolution of mixture spectra from ion-trap data-independent-acquisition tandem mass spectrometry. Anal. Chem. 82, 833–841 (2010).
Schubert, O.T. et al. The Mtb Proteome Library: a resource of assays to quantify the complete proteome of Mycobacterium tuberculosis. Cell Host Microbe 13, 602–612 (2013).
MacLean, B. et al. Effect of collision energy optimization on the measurement of peptides by selected reaction monitoring (SRM) mass spectrometry. Anal. Chem. 82, 10116–10124 (2010).
Zhang, Y., Ficarro, S.B., Li, S. & Marto, J.A. Optimized Orbitrap HCD for quantitative analysis of phosphopeptides. J. Am. Soc. Mass Spectrom. 20, 1425–1434 (2009).
Bereman, M.S. Tools for monitoring system suitability in LC-MS/MS–centric proteomic experiments. Proteomics 15, 891–902 (2015.
Bereman, M.S. et al. Implementation of statistical process control for proteomic experiments via LC-MS/MS. J. Am. Soc. Mass Spectrom. 25, 581–587 (2014).
Mallick, P. et al. Computational prediction of proteotypic peptides for quantitative proteomics. Nat. Biotechnol. 25, 125–131 (2007).
Stergachis, A.B., MacLean, B., Lee, K., Stamatoyannopoulos, J.A. & MacCoss, M.J. Rapid empirical discovery of optimal peptides for targeted proteomics. Nat. Methods 8, 1041–1043 (2011).
Acknowledgements
Financial support for this work was provided from US National Institutes of Health grants P41 GM103533, R01 GM103551, R21 CA192983 and U54 HG008097.
Author information
Authors and Affiliations
Contributions
J.D.E., B.M., R.J., Y.X. and M.J.M. developed and optimized the protocol. J.D.E. drafted the text of the manuscript. R.J. prepared samples and acquired data presented in ANTICIPATED RESULTS section.
Corresponding author
Ethics declarations
Competing interests
M.J.M. is a paid consultant for Thermo Fisher Scientific. Y.X. is an employee of Thermo Fisher Scientific. The MacCoss laboratory receives research support from Thermo Fisher Scientific.
Supplementary information
Supplementary Text and Figures
Supplementary Note and Supplementary Methods (PDF 746 kb)
Rights and permissions
About this article
Cite this article
Egertson, J., MacLean, B., Johnson, R. et al. Multiplexed peptide analysis using data-independent acquisition and Skyline. Nat Protoc 10, 887–903 (2015). https://doi.org/10.1038/nprot.2015.055
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2015.055
This article is cited by
-
Comparative proteomic analysis of seminal plasma exosomes in buffalo with high and low sperm motility
BMC Genomics (2023)
-
Q-RAI data-independent acquisition for lipidomic quantitative profiling
Scientific Reports (2023)
-
Utilizing Skyline to analyze lipidomics data containing liquid chromatography, ion mobility spectrometry and mass spectrometry dimensions
Nature Protocols (2022)
-
Dynamic FMR1 granule phase switch instructed by m6A modification contributes to maternal RNA decay
Nature Communications (2022)
-
Elamipretide effects on the skeletal muscle phosphoproteome in aged female mice
GeroScience (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.