Abstract
Two new antimycin antibiotics; that is antimycins A19 (1) and A20 (2), were isolated from a cultured broth of marine actinomycete Streptomyces antibioticus H74-18 together with antimycins A1a (3a) and A1b (3b), A2a (4), A3a (5a) and A3b (5b). Their structures were determined by spectroscopic methods in combination with X-ray diffraction. Antimycin A19 possessed a chiral acyl chain and an alkyl branch. The absolute configuration of chiral acyl chain in 1 was determined by X-ray diffraction analysis. Antimycin A20 (2) has the shortest and simplest acetoxy acyl chain in the antimycins family. All the antimycins (1–5) showed potential antifungal activities against Candida albicans with MIC of about 5–10 μg ml–1.
Similar content being viewed by others
Introduction
Most of the natural products with antibiotic activity have been produced by actinomycetes and fungi isolated from terrestrial environments. Actinomycetes have been the major source of new anti-infective agents, which include some of the most important and industrial antibiotic drugs.1 However, the discovery of completely novel metabolites from these microbes is becoming more and more difficult. In order to increase the discovery of drugs and lead compounds from natural products, we have focused on the isolation of the unique microorganisms from new, unexplored environments such as marine environment.
In the future, marine microbes may be a key source for commercially important bioactive compounds.2 Mangrove, being a unique inter-tidal ecosystem in tropics and subtropics, is ideally situated at the inter-phase between the terrestrial and marine environment and supports a rich and diverse aquatic and terrestrial microorganisms.3, 4 The mangrove environment is a valuable source for the isolation of antibiotic-producing actinomycetes.2, 5 Some antibiotics, including some specific and potent antifungal antibiotics, have been obtained from marine actinomycetes.6, 7, 8, 9, 10
Sediments of mangrove sites along the coast of the South China Sea have been a rich source of marine microorganism, especially actinomycetes.11, 12, 13, 14, 15, 16, 17 Our research group has isolated >2000 strains of actinomycetes from the sediment soils collected at several mangrove sites in the South China Sea, of which about 40 strains were found to show antifungal activity against Candida albicans by primary in vitro screening. Further characterization of antifungal compounds, which was produced by a cultured broth of Streptomyces antibioticus H74-18, led to the isolation of two new antibiotic compounds—antimycins A19 and A20 (Figure 1). In the report, we describe the taxonomy characteristics and fermentation of the antimycin-producing strain, and the isolation, structure determination and antifungal activities of antimycins A19 and A20.
Results and discussion
Strain H74-18 was isolated from a sediment soil collected at a mangrove zone in the South China Sea, Guangdong province, China. The vegetative mycelia grew abundantly on KIA agar (disaccharide iron medium), Glucose-gelatin-L-aspartic acid-agar, potato stock and Gaoshi No. 1 agar. The basal mycelia did not show fragmentation. The aerial mycelia has grayish white to dark gray color, and the basal mycelia has yellow to brownish red color. The conidiophore grown in aerial mycelia were curved, and each had about eight spores per conidiophore. The spores were cylindrical in shape, about 2 × 0.6 mm2 in size, and had a smooth surface (Figure 2). However, both types of mycelia grew sparsely in white color on Czapek Dox Agar and starch media. The pigments generated by the strains easily diffused on Gaoshi No. 1 agar, partly on potato stock medium, did not diffuse on glucose-aspartic acid-agar and starch agar media. No pigment was generated on the other media. The strains can utilize the six types of carbon resources, liquidize the gelatin, lyse starch, but cannot grow on cellose, cannot peptonize and liquidize milk and cannot generate black pigment. Chemical analysis revealed that the lysis solution of the whole cells from the H74-18 contained L-DAP, glycine, alanine, L-aspartic acid, ribose, glucose and non-characteristic monosaccharose. The chemical composition of strain cell wall belonged to type I, inconsistent with chemotaxonomic characteristic of genus Streptomyces. 16S rDNA sequence and phylogenetic tree analyses showed that the strain was most closely related to S. antibioticus, sharing 16S rDNA similarity value of 99.9%. Based on the taxonomic properties described above, the strain H74-18 was considered to be S. antibioticus.
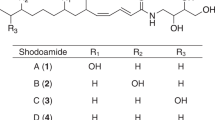
Compound 1 was obtained as colorless plate crystal from a petroleum ether–acetone solvent system. High-resolution ESI-MS give quasi-molecular ion peak m/z 549.2729 [M+H]+, suggesting its molecular formula C28H40N2O9 (calcd. for C28H41N2O9, 549.2807). Because of the small amount of compound 1 and the available crystals, single crystal X-ray diffraction analysis was undertaken for the determination of its structure. The result indicated that it is a member of antimycins. The basic skeleton of compound 1 was composed of a benzyl ring (A), a nine-member dilactone ring (B) and two side chains (Figures 3 and 4). The dihedral angle between rings A and B was 48.6° (Figure 5). Because of the longer side chains attached at two adjacent chiral carbons (C7 and C8), the rotation around single bonds C8/C12 and C7/O led to the severe derivation from normal value of the temperature factors of atoms on the side chains. The strong thermal vibration of C-8′ groups connected to nitrogen atom also resulted in the severe derivation from normal value of the temperature factors of atoms on the moieties. The arrangement of molecules in crystal belonged to the first type of space group, suggesting that this sample is optical active. The strong intramolecular hydrogen bond (D=2.543 ˚) linked phenolic OH with o-carbonyl O atom in the molecule. The inter-molecular hydrogen bond (D=2.801 ˚) also formed between N and O atoms from two different formamide. The S absolute configuration of the chiral center in the acyl side chain was determined by X-ray diffraction analysis due to the previous determination of other chiral centers in the molecule. This is the first antimycin with explicit absolute configuration of chiral carbons in the acyl side chains. This new antibiotic compound had the chemical name (2S,3R,6R,7S,8S,2′R)-3-(3-formamido-2-hydroxybenzamido)-8-pentyl-2,6-dimethyl-4,9-dioxo-1,5-dioxonan-7-yl-2′,3′-dimethyl propionic ester, and was trivially named as antimycin A19.
Compound 2 has the physico-chemical properties as follows: ESI-MS m/z: 505.2 [M–H]−, 529.2 [M+Na]+ (see Figure 5), HR-ESI-MS m/z: 505.2185 [M–H]− (calcd. for C25H33N2O9, 505.2192). 1H NMR (400 MHz, CDCl3) data (see Table 1) were very similar to those of antimycins. Because of a very small amount of 2, 13C NMR and 2D data were not successfully obtained. The existence of proton signal at δ 2.13 (3H, s) and the absence of the other proton signals from acyl group side chains easily seen in other antimycins revealed that the acyl group in 2 was an acetoxy. So, 2 was elucidated to be (2S,3R,6R,7S,8S)-3-(3-formamido-2-hydroxybenzamido)-8-hexyl-2,6-dimethyl-4,9-dioxo-1,5-dioxonan-7-yl acetate, named antimycin A20. Compound 2 is a member of antimycins having the shortest and simplest acetoxy as acyl side chain.
Samples 3∼5 were a HPLC single-peak samples prepared with HPLC from a crystal mixture. We have not found a HPLC eluting system to separate it into two peaks on analytical HPLC ODS column. So, 3∼5 were directly used to measure its ESI-MS, 1D and 2D NMR data. However, The NMR data indicated that both 3 and 5 are the mixture of antimycin A1a/A1b and A3a/A3b, respectively. Most of the 1H and 13C signals from basic skeleton of dilactone ring and benzyl ring of two components in both samples 3 and 5 had completely overlapped on the spectra. Two different acyl groups could be analyzed due to their discrete 1H and 13C signals, as seen in Hosotani et al.18 Sample 4 was a single compound antimycin A2a on the basis of its 1H and 13C NMR characteristics.
A paper disc method with C. albican was used to guide the fractionation and purification of bioactive molecules. Antifungal activities against C. albican was measured using a 96-well plate assay with final concentration of compounds of 20, 10, 5, 2.5, 1.25 and 0.75 μg ml–1. The assay plates were incubated for about 20 h, with DMSO as the negative control and clotrimazole as the positive control. The clear wells with minimum concentration of compounds were counted for the MIC concentration. Samples 1–5 exhibited different MICs at 5 (1), 10 (2), 10 (3a and 3b), 5 (4), 10 (5a and 5b) μg ml–1, respectively. Their MICs against Cryptococcus neoformans and Aspergillus niger were higher than 20 μg ml–1 by the same method.
Experimental procedure
General experimental procedures
The morphological properties were observed with a JEOL JSM-5600 scanning electron microscope (JEOL Ltd., Tokyo, Japan). UV was recorded on a Jasco V-550 UV/vis spectrophotometer (Jasco International, Tokyo, Japan). IR was run on a Jasco FI/IR plus Fourier transform spectrometer (Jasco International). NMR spectra were recorded at 25 °C on a Bruker 400 MHz spectrometer (Bruker Corporation, Fallanden, Switzerland) with residual CDCl3 peak as reference. LR-ESI-MS spectra were recorded on a Finnigan LCQ Advantage MAX mass spectrometer spectrometer (Thermo Electron Corporation, San Jose, CA, USA). HR-ESI-MS data were recorded on an AB4000 Q Trap mass spectrometer (Agilent, Santa Clara, CA, USA). Preparative HPLC separation was performed on Gilson 306 (pump) (Gilson Inc., Villiers Le Bel, France) equipped with an UV/VIS-152 detector and Waters SunFire Prep C18 Φ 19 × 150 mm2 (Waters Corporation, Milford, MA, USA). All solvents used for chromatography, UV and MS were Lab-Scan HPLC grade (POCH S.A., Gliwice, Poland), and the H2O was filtered with Millipore Milli-Q PF (Millipore Corporation, Billerica, MA, USA). Strain C. albican in paraffinic oil suspension solution of the spore was purchased from Guangdong Institute for Drug Control. Strains C. neoformans and A. niger were supplied by the Department of Microbiology, Medical College, Jinan University.
Fermentation
A stock culture of Streptomyces sp. H74-18 was inoculated into a large test tube containing 10 ml of a seed. Strain H74-18 grew on Gaoshi No. 1 agar media at 28 °C for 14 days. The medium consisted of glucose 40.0 g, peptone 10.0 g, agar 20.0 g in 1000 ml distilled water with 0.05 mg ml–1 chloromycetin. The fermentation was carried out on a reciprocal shaker at 28 °C for 2 days. One milliliter of the first seed culture was transferred into each of eight 500 ml Erlenmeyer flasks containing 100 ml of the same seed medium and incubated on a rotary shaker at 28 °C for 3 days. Then, 2000 ml of the second seed culture was transferred into a 30-l jar fermenter containing 20 l of production medium that consisted of the same media mentioned above.
Extract and isolation
The whole culture broth (20 l) was filtered. The mycelia was soaked and extracted with acetone and condensed by rotary evaporation under reduced pressure. The aqueous extract was suspended in distilled water and partitioned with ethyl acetate (EtOAc), and the organic layer was concentrated in vacuo to afford the crude antifungal material (50.0 g), which was chromatographed over a silica gel column (1000 g). Active fractions eluted with petroleum ether–ethyl acetate (5:1) were concentrated to yield an active crude subfraction (1.07 g), which was applied to another silica gel column and eluted with petroleum ether–acetone (7:1). A crude crystal sample (25 mg) with potent antifungal activity was obtained from some fractions. The crystal sample contained a few morphological types of crystals including plate, columnar crystal and acicular crystal. LC-ESI-MS (positive) on Agilent Eclipse Plus and RP-C18 (5 μm) column (4.6 mm × 250 mm; Welch Material Inc., Ellicott, MD, USA), eluting with methanol-0.5% formic acid distilled water (80:20) solvent system and flowing rate at 1 ml min–1, revealed that it is a mixture with five major peaks with ion peaks at m/z 543.5, 557.7, 557.7, 571.7 and 571.7, respectively and the corresponding retention times at 6.33, 7.98, 8.67, 11.37 and 12.31. A single plate-like crystal (1) was picked up for X-ray diffraction analysis. The rest of the crystal sample (23 mg) and residue from the mother solution were applied onto an ODS HPLC (i.d. 19 × 150 mm2, Waters SunFire Prep C18 column), which was eluted with methanol- 0.5% formic acid distilled water (80:20) with a detection wavelength at 240 nm to afford four colorless powders; that is antimycin A19 (2, 1.1 mg), antimycins 1a and 1b (3, 4.2 mg), antimycin 2a (4, 3.5 mg) and antimycins 3a and 3b (5, 5.0 mg), respectively.
X-ray diffraction
The crystallographic analysis of compound 1 was described below: formula C28H40N2O9; Mr 548.62; monoclinic system; space group: P21; a=12.553 (1) Å, b=8.026 (1) Å, c=15.708 (1) Å; β=92.06 (1) Å; V=1581.1 (7) Å3; Z=2; crystal dimensions 0.10 × 0.15 × 0.20 mm3, calculated density 1.152 g cm–3. Diffraction intensity data were collected by using a MAC DIP-2030K diffractometer (Mac Science, Tokyo, Japan) to a maximum 2θ value of 50.0° with graphite-monochromated MoKα radiation. A total of 6868 independent reflections were collected, of which 2897 were observed (∣F∣2⩾4σ∣F∣2). The crystal structures were solved by direct methods using Shelxs-97 (University of Göttingen, Göttingen, Germany). In the structure refinements, non-H atoms were refined anisotropically. H atoms bonded to carbons were placed on the geometrically ideal positions by the “ride on” method. H atoms bonded to oxygen were located by the difference Fourier method and were included in the calculation of structure factors with isotropic temperature factors. The final R1=0.0621 and wR2=0.1473, S=0.950.
Antifungal assay
Antifungal activities were determined using MIC method in 96-well plates with compounds diluted in DMSO. C. albicans, C. neoformans and A. niger were grown in modified Martin broth for 12 h and dispensed into 96-well plates, 200 μl per well. Compounds were added to 96-well plates at final concentrations of 20, 10, 5, 2.5, 1.25 and 0.625 μg ml–1. Plates were incubated for 20 h, with DMSO as negative control. Modified Martin broth was composed of glucose 20 g, peptone 5 g, Yeast extract 4 g, K2HPO4•7H2O 0.63 g, MgSO4•7H2O 1.8 g in 1000 ml distilled water. MIC was determined as the concentration of compound that totally inhibited growth of C. albicans.
Supporting information on CIF data (CCDC 827453) for the crystal structure of new compound 1 and NMR spectra of compound 2 are available from the corresponding author or free of charge via the Internet at the IUCr electronic archives.
References
Bernan, V. S., Greenstein, M. & Carter, G. T. Mining marine microorganisms as a source of new antimicrobials and antifungals. Curr. Med. Chem. Anti-Infect. Agents 3, 181–195 (2004).
Das, S., Lyla, P. S. & Ajmal, K. S. Marine microbial diversity and ecology: importance and future perspectives. Curr. Sci. 90, 1325–1335 (2006).
Sahoo, K. & Dhal, N. K. Potential microbial diversity in mangrove ecosystems: a review. Indian J. Mar. Sci. 38, 249–256 (2009).
Dias, A. C. F. et al. Diversity and biotechnological potential of culturable bacteria from Brazilian mangrove sediment. World J. Microbiol. Biotechnol. 25, 1305–1311 (2009).
Rathna, K. R. R. & Chandrika, V. Effect of different media for isolation, growth and maintenance of actinomycetes from mangrove sediments. Indian J. Mar. Sci. 22, 297–299 (1993).
Colwell, R. R. & Hill, R. T. Microbial diversity. In Diversity of Oceanic Life: An Evaluative Review (ed. Peterson, M. N. A.) 100–106 The Center for Strategic and International Studies, Washington, DC, 1992.
Kokare, C. R., Mahadik, K. R., Kadam, S. S. & Chopade, B. A. Isolation, characterization and antimicrobial activity of marine halophilic Actinopolyspora species AH1 from the west coast of India. Curr. Sci. 86, 593–597 (2004).
Zhou, M. Y. & Zheng, Z. C. Identification of marine actinomycetes S-216 strain and its biosynthetic conditions of antifungal antibiotic. J. Xiamen Univ. Nat. Sci. 37, 109–114 (1998).
David, J.N. & Russell, T.H. New drugs from marine microbes: the tide is turning. J. Ind. Microbiol. Biotechnol. 33, 539–544 (2006).
Rathna, K. R. & Chandrika, V. Microbial production of antibiotics from mangrove ecosystem. CMFRI Spl. Publ. 61, 117–122 (1995).
Hong, K. et al. Actinomycetes for marine drug discovery isolated from mangrove soils and plants in China. Mar. Drugs 7, 24–44 (2009).
Xu, Y. J., Chen, S. Z., Luo, N. X., Kong, J. & Huang, D. L. Isolation of actinomycetes from mangrove in Guangxi and extraction of its genomic DNA. Anhui Nongye Kexue 37, 15155–15156 (2009).
Xiao, J. et al. Isolation of mangrove actinomycetes and their antagonistic activities. Yingyong Yu Huanjing Shengwu Xuebao 14, 244–248 (2008).
Xu, J. et al. Streptomyces xiamenensis sp. nov., isolated from mangrove sediment. Int. J. System. Evolut. Microbiol. 59, 472–476 (2009).
Huang, D., Chen, S. & Xu, Y. Method of isolation and identification for genomic DNA extraction from soil of mangrove forest actinomycetes in guangxi. Guangxi Yixue 30, 1657–1659 (2008).
Huang, H. et al. Micromonospora rifamycinica sp. nov., a novel actinomycete from mangrove sediment. Int. J. System. Evolut. Microbiol. 58, 17–20 (2008).
Jiang, X., Liang, X. & Cao, J. Selection of antibiotic actinomyces from mangrove forest ecosystem in Futian, Shenzhen. Zhongguo Haiyang Daxue Xuebao (Ziran Kexueban) 36, 601–605 (2006).
Hosotani, N., Kumagai, K., Nakagawa, H., Shimatani, T. & Saji, I. Antimycins A10_A16, seven new antimycin antibiotics produced by Streptomyces spp. SPA-10191 and SPA-8893. J. Antibiot. 58, 460–467 (2005).
Acknowledgements
This work was supported by the Grant (No. 30873199) of National Natural Science Foundation of China, and Program for New Century Excellent Talents in University (NCET-08-0612). We thank Professor Lu Yang’ research group in Institute of Materia Medica, Chinese Academy of Medical Science (Beijing) for analysis of X-ray diffraction. All NMR data and HR/LR-ESI-MS were measured in Institute of Traditional Chinese Medicine and Natural Products, College of Pharmacy, Jinan University.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Xu, LY., Quan, XS., Wang, C. et al. Antimycins A19 and A20, two new antimycins produced by marine actinomycete Streptomyces antibioticus H74-18. J Antibiot 64, 661–665 (2011). https://doi.org/10.1038/ja.2011.65
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ja.2011.65
Keywords
This article is cited by
-
The microbiome of a shell mound: ancient anthropogenic waste as a source of Streptomyces degrading recalcitrant polysaccharides
World Journal of Microbiology and Biotechnology (2021)
-
Antimicrobial compounds from marine actinomycetes
Archives of Pharmacal Research (2020)
-
Structural Elucidation and Identification of 2-Hydroxy Benzoic Acid: An Antibacterial and Cytotoxic Compound from Streptomyces sp. VITHM1 Isolated from Marine Sediment Sample of Alappuzha Beach, Kerala, India
Arabian Journal for Science and Engineering (2018)
-
Opantimycin A, a new metabolite isolated from Streptomyces sp. RK88-1355
The Journal of Antibiotics (2017)
-
Endophytic Streptomyces sp. AC35, a producer of bioactive isoflavone aglycones and antimycins
Journal of Industrial Microbiology and Biotechnology (2016)