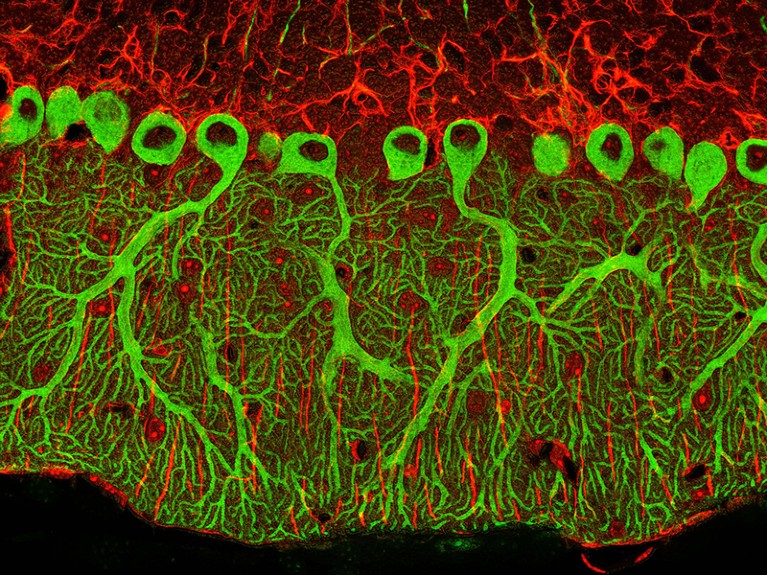
An electron micrograph of cerebellum tissue. Nerve cells are bright green.Credit: Thomas Deerinck, NCMIR/SPL
Researchers have created the first complete atlas of cells in the mouse brain, revealing an incredible 5,322 cell types1,2. The work, spanning ten papers — nine of them published by Nature on 13 December — marks the final outcome of the flagship US project, the Brain Research through Advancing Innovative Neurotechnologies (BRAIN) Initiative Cell Census Network (BICCN).
Funded by the National Institutes of Health to the tune of US$375 million, the BICCN was launched in 2017 as a five-year collaboration between 250 scientists at 45 institutions across three continents.
Cellular atlases of the entire mouse brain
Its goal was to use single-cell and spatial-genomics technologies to define and localize brain cell types across humans, non-human primates and mice. The ultimate aim is to improve understanding of the cellular mechanisms behind brain disorders, a leading cause of death and disability worldwide. In October, BICCN researchers generated the largest atlas of human brain cells yet, with more than 3,000 cell types3.
The BICCN mouse-brain atlas, which is open access, is also intended to serve as a reference for scientists studying brain disorders and brain evolution across species. It is not the only one of its kind. Several mega brain projects, each trying to make their own reference atlas or reference model, are under way around the world.
These include Japan’s Brain/MINDS project to map the marmoset (Callithrix jacchus) brain4, China’s Brain Project, focusing on macaque monkeys (Macaca spp.)5 and the Korea Brain Initiative, which aims to develop specialized brain maps for both mice and humans6.
Scientists at the European Union’s Human Brain Project (HBP), which was based in Geneva, Switzerland, and ended in September, have also created the Human Brain Atlas. This is available on the HBP’s open platform EBRAINS. The atlas uses post-mortem brain-imaging data and depicts the multilevel organization of the brain, from its cellular and molecular architecture to its functional modules and neural connections.
The Swiss Blue Brain Project, which will wrap up in 2024 after nearly 20 years, will also release a digital model of the mouse brain, based on imaging data, with detailed mapping of neurons and connectivity circuits.
Each of these projects is important in its own right, but as they progress — some have been going more than ten years — there is a need for better communication between them. Several of the projects are using similar or identical technologies. It makes sense for the teams to liaise more closely, at the very least to begin a discusson on how to establish shared data standards, which they have not yet done.
Closer liaison
The BICCN atlas was built using a technology called high-resolution single-cell transcriptomics, which defines cells by the genes that they express and uses imaging technologies to map the genes’ spatial distribution. The atlas details how many brain cell types there are, as well as their proportions and spatial arrangement in the brain.
In 2021, BICCN researchers mapped these cell types in around 1% of the mouse brain. Advancements in these genomic technologies allowed the team to catalogue 90% of cell types in the adult mouse brain in two years. The NIH has launched another five-year project called the BRAIN Initiative Cell Atlas Network, with the aim of scaling up the technologies to map the brain cell types of humans and non-human primates, as well as the mouse brain, during the organ’s development.
Read the paper: The molecular cytoarchitecture of the adult mouse brain
These same technologies underpin the China Brain Project, which is being funded with 3.2 billion Chinese yuan (US$446 million). As with the NIH study, its goal is to map macaque brain cell types using genomic technologies — it has already used these tools to generate an atlas of 143 regions in the macaque brain cortex and has identified 264 cortical cell types7.
Meanwhile, the Korea Brain Initiative, which is running from 2016 to 2026, intends to construct maps at several scales, in both the prefrontal cortex and the basal ganglia. Its researchers are using nano- and mesoscale mapping and single-cell transcriptome analysis, the same technologies being deployed by the NIH and the China Brain Project.
At a minimum, the data, models and code need to be open. Even then, very large data sets such as these still pose a challenge to reproducibility. That is why a standardized framework for data collection and analysis, including definitions for types of cell clusters, as well as unified cell classifications and names across species, will eventually be needed.
Such standardization will be an important goal, albeit a longer-term one. An interim step must be to begin a discussion on shared data standards. Setting these will help to unlock the full potential of these brain maps, allowing the groups to better understand and build on each other’s findings.


 Cellular atlases of the entire mouse brain
Cellular atlases of the entire mouse brain
 Read the paper: A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain
Read the paper: A high-resolution transcriptomic and spatial atlas of cell types in the whole mouse brain
 Read the paper: The molecular cytoarchitecture of the adult mouse brain
Read the paper: The molecular cytoarchitecture of the adult mouse brain
 Europe spent €600 million to recreate the human brain in a computer. How did it go?
Europe spent €600 million to recreate the human brain in a computer. How did it go?
 How the world’s biggest brain maps could transform neuroscience
How the world’s biggest brain maps could transform neuroscience