Abstract
Increasing autophagy is beneficial for curing hepatocellular carcinoma (HCC). Damage-regulated autophagy modulator (DRAM) was recently reported to induce apoptosis by mediating autophagy. However, the effects of DRAM-mediated autophagy on apoptosis in HCC cells remain unclear. In this study, normal hepatocytes (7702) and HCC cell lines (HepG2, Hep3B and Huh7) were starved for 48 h. Starvation induced apoptosis and autophagy in all cell lines. We determined that starvation also induced DRAM expression and DRAM-mediated autophagy in both normal hepatocytes and HCC cells. However, DRAM-mediated autophagy was involved in apoptosis in normal hepatocytes but not in HCC cells, suggesting that DRAM-mediated autophagy fails to induce apoptosis in hepatoma in response to starvation. Immunoblot and immunofluorescence assays demonstrated that DRAM translocated to mitochondria and induced mitophagy, which led to apoptosis in 7702 cells. In HCC cells, starvation also activated the phosphatidylinositol 3-kinase (PI3K)/AKT pathway, which blocks the translocation of DRAM to mitochondria through the binding of p-AKT to DRAM in the cytoplasm. Inactivation of the PI3K/AKT pathway rescued DRAM translocation to mitochondria; subsequently, mitochondrial DRAM induced apoptosis in HCC cells by mediating mitophagy. Our findings open new avenues for the investigation of the mechanisms of DRAM-mediated autophagy and suggest that promoting DRAM-mediated autophagy together with PI3K/AKT inhibition might be more effective for autophagy-based therapy in hepatoma.
Similar content being viewed by others
Main
Hepatocellular carcinoma (HCC) is the fifth most common cancer worldwide and the third most common cause of cancer deaths.1 HCC is a common malignant tumor and causes over 100 000 HCC deaths per year in China.2 Recently, studies have indicated that increasing autophagy is beneficial for curing HCC.3, 4, 5, 6 Although under conditions of metabolic stress, induction of autophagy can provide nutrients and energy required for cell viability.7, 8 Autophagy has emerged as a key regulator of death pathways.9, 10, 11 The mechanisms of autophagy-dependent cell death might be partially associated with the activation of caspase-dependent pathways.9, 12 Moreover, we recently reported that the induction of damage-regulated autophagy modulator (DRAM)-mediated autophagy induces apoptotic cell death in hepatoma cells.13
p53 has been reported to induce DRAM-mediated autophagy, which is a pro-apoptotic factor.14 p73 has also been identified to induce DRAM expression when p53 is deficient; however, p73-induced DRAM is not involved in the induction of autophagy and apoptosis.15 DRAM contains a putative signal peptide for targeting to the endoplasmic reticulum (ER) and six hydrophobic potential transmembrane regions.14 DRAM can localize to many different subcellular sites.16 Although the relationship between DRAM function and subcellular localization remains unclear, our previous study suggests that mitochondrial DRAM induces apoptosis in hepatoma cells by mediating mitophagy.13 However, the effect of inducing DRAM-mediated autophagy in cancer cells has not been well studied until now.
A clear link has been established between the phosphatidylinositol 3-kinase (PI3K)/AKT pathway and the pathogenesis of HCC.17, 18 PI3Ks primarily phosphorylate phosphatidylinositol-4,5-bisphosphate, generating the lipid second messenger phosphatidylinositol-3,4,5-trisphosphate (PIP3).19 AKT, a key factor in the PI3K pathway, can be recruited to the membrane via binding to PIP3 through its pleckstrin homology domains and is fully activated following phosphorylation.20 Activation of AKT can predict poor prognosis in HCC.21 Constitutive activation of the PI3K/AKT signaling pathway often causes cells to proliferate in an uncontrolled manner. The anti-apoptotic activity caused by AKT activation has been suggested to depend on its translocation from the cytosol to the mitochondria, where it inhibits opening of the permeability transition pore to maintain mitochondrial integrity.22
In this study, we used a normal liver cell line (7702) and three HCC cell lines (HepG2, Hep3B and Huh7) to detect the effect of DRAM-mediated autophagy on apoptosis induced by serum deprivation. We also assessed whether DRAM-mediated autophagy and the PI3K/AKT pathway engage in crosstalk that affects apoptosis.
Results
Autophagy is involved in starvation-induced apoptosis in 7702 cells but not HCC cell lines
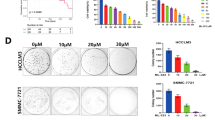
A normal liver cell line (7702 (wild-type p53)) and HCC cell lines (HepG2 (wild-type p53), Hep3B (p53 null) and Huh7 (p53 A220G)) were grown in serum-free medium for 48 h. Using M30 immunoreactivity and calcein AM/propidium iodide (PI) to detect early and late apoptosis, we observed that starvation-induced apoptosis in all cell lines (Figures 1a and b). Moreover, starvation induced higher levels of apoptosis in 7702 cells than in HepG2, Hep3B and Huh7 cells (Figures 1a and b). Immunoblot and flow cytometry assays also showed that the level of apoptosis was higher in 7702 than in three HCC cell lines in response to starvation (Figure 1c and Supplementary Figure 1a). The apoptosis level of HepG2, Hep3B and Huh7 cells was not significantly different in response to starvation (Figures 1a–c and Supplementary Figure 1a). Serum deprivation also induced autophagy in all cell lines (Figure 1c); however, inhibition of autophagy via Bafilomycin A1 (BafA 1) or NH4Cl decreased the apoptosis of 7702 cells but not the three HCC cell lines (Figures 1d–g and Supplementary Figures 1b–d). Thus, starvation-induced autophagy induces apoptosis, which causes 7702 cells to become more sensitive to apoptosis.
Starvation-induced autophagy involves apoptosis in 7702 cells but not in three HCC cell lines. The 7702 (wild-type p53) and HCC cell lines (HepG2 (wild-type p53), Hep3B (p53 null) and Huh7 (p53 A220G)) were starved (sta) for 48 h with or without pre-treatment with BafA 1 to inhibit autophagy. (a) M30 immunoreactivity and calcein AM/PI assays were used to detect apoptosis in all cell lines in response to starvation. M30 and PI staining (red) indicated early and late apoptotic cell death, respectively. Nuclei were stained with DAPI. Representative images of cells were obtained with a fluorescence microscope at × 40 magnification. (b) Quantitation of M30-positive and PI-positive cells observed in (a). (c) Autophagy and apoptosis were detected in cells in response to starvation for 48 h via immunoblot analysis with the indicated antibodies (anti-LC3 and p62 antibodies for detecting autophagy; anti-PARP antibody for detecting apoptosis). (d and e) An immunoblot assay was used to detect the effect of autophagy inhibition via BafA 1 on starvation-induced apoptosis in cells. (f and g) Quantification of M30-positive and PI-positive cells starved for 48 h with or without autophagy inhibition using BafA 1. The data are presented as the mean±S.D. of three independent experiments in (b), (f) and (g)
DRAM-mediated autophagy is a critical inducer of apoptosis in 7702 cells but not in HepG2, Hep3B or Huh7 cells in response to starvation
Crighton et al.14 and our previous study have demonstrated that DRAM-mediated autophagy induces apoptosis.13 In this study, we observed that DRAM mRNA and protein levels were increased in 7702 and the three HCC cell lines in response to starvation (Figures 2a and b). All cell lines were transfected with GFP-microtubule-associated protein light chain 3 (LC3) plasmids, and an immunofluorescence assay determined that starvation induced the development of GFP-LC3 puncta (Figure 2c). In addition, siRNA-mediated knockdown of DRAM significantly reduced the ratio of GFP-LC3 puncta-positive cells (Figures 2c and d). An immunoblot assay also showed that siRNA-mediated DRAM knockdown reduced the level of autophagy (Figure 3a). DRAM siRNA effectively knocked down DRAM expression in all cell lines (Figure 2e), and control siRNA had no effect on starvation-induced autophagy (Figures 2c and d). Thus, our data suggest that DRAM is involved in starvation-induced autophagy in normal hepatocytes and HCC cells. Moreover, immunoblot and immunofluorescence assays showed that knockdown of DRAM via siRNA reduced apoptosis in 7702 cells but not in the three HCC cell lines in response to starvation (Figures 3a–c). Taken together, these data reveal that DRAM-mediated autophagy contributes to starvation-induced apoptosis in normal hepatocytes, but not in HCC cells.
Starvation induces DRAM-mediated autophagy in 7702, HepG2, Hep3B and Huh7 cells. (a and b) The 7702, HepG2, Hep3B and Huh7 cell lines were starved for 48 h, and real-time PCR (a) and immunoblot (b) assays were used to detect DRAM expression. The immunoblot assay was also used to detect the expression of p53, p73 and LC3I/II (b). (c) The 7702, HepG2, Hep3B and Huh7 cell lines were starved (sta) for 48 h with or without pre-transfection with DRAM siRNA (DRAM si) or control siRNA (control si). Confocal microscopy was used to detect the formation of GFP-LC3 puncta. Magnification, × 40. (d) Quantification of cells with >5 GFP-LC3 puncta. (a and d) The data are presented as the mean±S.D. of three independent experiments. (e) The effect of DRAM siRNA on DRAM expression in cells with or without starvation for 48 h was determined using an immunoblot assay
DRAM-mediated autophagy in response to starvation induces apoptosis only in 7702 cells and not in three HCC cell lines. The 7702, HepG2, Hep3B and Huh7 cell lines were transfected with DRAM siRNA (DRAM si) and were then starved (sta) for 48 h. (a) An immunoblot assay was used to detect LC3 I/II and cleaved PARP fragment (p85). (b and c) Quantification of starvation-induced M30-positive and PI-positive cells with or without DRAM knockdown using DRAM si. The data are presented as the mean±S.D. of three independent experiments
p53 has a critical role in inducing DRAM-mediated autophagy in 7702 and HepG2 cells in response to starvation
DRAM-mediated autophagy is induced in both 7702 and HepG2 cells, which are both wild-type for p53. p53 knockdown by siRNA completely blocked DRAM expression and significantly reduced the level of autophagy (Figure 4a); p53 siRNA treatment did not affect the expression of p73 (data not shown), which highlights the key role of wild-type p53 in inducing DRAM-mediated autophagy in 7702 and HepG2 cells in response to starvation. In 7702 cells, siRNA-induced knockdown of DRAM significantly reduced starvation-induced apoptosis; the effect of siRNA knockdown of both p53 and DRAM was equivalent to the effect of siRNA knockdown of p53 alone with respect to apoptosis reduction, suggesting that DRAM-mediated autophagic apoptosis is a downstream effect of activated p53 in 7702 cells (Figure 4b). In HepG2 cells, siRNA-induced p53 knockdown almost completely blocked starvation-induced apoptosis; however, DRAM knockdown by siRNA did not affect starvation-induced apoptosis, suggesting that the apoptosis-inducing role of p53 is not impaired in hepatoma cells with wild-type p53, although p53-induced DRAM cannot induce apoptosis by mediating autophagy.
p53 is critical for the induction of DRAM-mediated autophagy in 7702 and HepG2 cells expressing wild-type p53. (a) The 7702 and HepG2 cell lines were transfected with p53 siRNA (p53 si) and were then starved (sta) for 48 h. An immunoblot assay was used to detect the effect of p53 knockdown by p53 si on the expression of DRAM, LC3 I/II, p62 and the cleaved PARP fragment (p85). (b) The 7702 and HepG2 cell lines were transfected with DRAM siRNA (DRAM si) or p53 si, or co-transfected with DRAM si and p53 si and were then starved for 48 h. M30 immunoreactivity (red) was used to detect the effect of siRNA-induced DRAM or p53 knockdown or co-knockdown of DRAM and p53 on apoptosis. Representative immunofluorescence images of cells were obtained with a fluorescence microscope at × 40 magnification. Nuclei were stained with DAPI
p73 has a critical role in starvation-induced DRAM-mediated autophagy in Hep3B (p53 null) and Huh7 (p53A220G) cells
In Hep3B cells with null p53 and Huh7 cells with an A220G mutation in p53, we determined that starvation induced p73 expression (Figure 2b). p73 knockdown using siRNA completely blocked DRAM expression and significantly decreased starvation-induced autophagy, suggesting that DRAM-mediated autophagy can be induced in a p73-dependent manner in Hep3B and Huh7 cells (Figure 5a). p73 is a member of the p53 family and shares a similar structure with p53, and we determined that p73 expression contributed to apoptosis in Hep3B and Huh7 cells (Figure 5b). However, DRAM siRNA did not affect the level of apoptosis in Hep3B and Huh7 cells in response to starvation (Figure 5b). These results suggest that the apoptosis-inducing role of p73 is not impaired in Hep3B or Huh7 cells; however, p73-induced DRAM cannot induce apoptosis by mediating autophagy.
Both starvation-induced p73 and rAd-p53-induced p53 overexpression can induce DRAM-mediated autophagy in Hep3B and Huh7 cells. (a) Hep3B and Huh7 cells were transfected with p73 siRNA (p73 si) and were then starved (sta) for 48 h. An immunoblot assay was used to detect the effect of p73 knockdown by siRNA on the expression of DRAM, LC3 I/II, p62 and cleaved PARP fragment (p85). (b) Hep3B and Huh7 cells were transfected with DRAM siRNA (DRAM si) or p73 si, or co-transfected with DRAM si and p73 si; the cells were then starved for 48 h. Starvation-induced M30-positive cells were quantified. The data are presented as the mean±S.D. of three independent experiments. (c) Hep3B and Huh7 cells were infected with rAd-p53 and were then starved for 48 h. An immunoblot assay was used to detect the effect of p53 overexpression on the expression of p73, DRAM, LC3 I/II and cleaved PARP fragment (p85). (d) Hep3B and Huh7 cells were infected with rAd-p53 with or without pre-treatment with DRAM siRNA and subsequently starved for 48 h. An immunoblot assay was used to detect the effect of DRAM knockdown via siRNA on autophagy. (e) rAd-p53-infected Hep3B and Huh7 cells were pre-treated with DRAM siRNA and were then starved for 48 h. M30 immunoreactivity (red) was used to detect the effect of siRNA-mediated DRAM knockdown on p53 overexpression-induced apoptosis. Nuclei were stained with DAPI. Representative immunofluorescence images of cells were obtained with a fluorescence microscope at × 40 magnification
Although p53 overexpression induces DRAM-mediated autophagy, the autophagy mediated by DRAM does not induce apoptosis in Hep3B or Huh7 cells
It has been reported that p73-dependent DRAM expression does not induce apoptosis,15 and our previous results also showed that p73-dependent DRAM-mediated autophagy was not involved in apoptosis. We investigated whether p53 overexpression following infection with rAd-p53 induced DRAM-mediated autophagic apoptosis in Hep3B and Huh7 cells. Our results revealed that p53 overexpression in these cells induced DRAM expression and DRAM-mediated autophagy in a p73-independent manner (Figures 5c and d). p53 overexpression induced apoptosis in Hep3B and Huh7 cells (Figures 5c and e and Supplementary Figures 2a and b); however, p53 overexpression-induced DRAM did not induce autophagic apoptosis (Figure 5e and Supplementary Figures 2c and d). As expected, rAd-p53 infection induced apoptosis in Hep3B and Huh7 cells, suggesting that p53 also has a role in inducing apoptosis (Figures 5c and e). Together with our previous results, these data demonstrate that the failure of DRAM-mediated autophagy to induce apoptosis in the three HCC cell lines is not caused by impairment in p53 or p73 function.
The starvation-activated PI3K/AKT pathway inhibits the induction of apoptosis by DRAM-mediated autophagy in HCC cell lines but not in 7702 cells
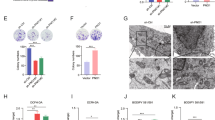
The PI3K/AKT signaling pathway is constitutively activated in HCC cells and other cancer cells.19 Using immunoblot assays, we observed that starvation induced the phosphorylation of PI3K and AKT in the three HCC cell lines but not in 7702 cells (Figure 6a). Inhibition of the PI3K/AKT pathway via PI3K siRNA significantly increased apoptosis in the three HCC cell lines but not in 7702 cells (Figure 6b). As activated PI3K/AKT consistently generates survival signals and protects cancer cells from apoptosis,19 we attempted to determine whether activation of the PI3K/AKT pathway could inhibit the effect of DRAM-mediated autophagy on the induction of apoptosis in the three HCC cell lines. Immunoblot analysis showed that blocking the PI3K/AKT pathway with PI3K siRNA or LY294002 did not affect the autophagy level or the expression of p53, p73 or DRAM in the three HCC cell lines (Figure 6c and Supplementary Figure 3). Interestingly, co-transfection of both PI3K and DRAM siRNAs into the three HCC cell lines significantly decreased apoptosis compared with transfection of DRAM siRNA alone (Figure 6d). Moreover, our results revealed that the increased level of apoptosis in the three HCC cell lines treated with PI3K siRNA is almost identical to the level of apoptosis in 7702 cells following starvation (Figure 6e). Taken together, our results suggest that activation of PI3K/AKT inhibits the induction of apoptosis triggered by DRAM-mediated autophagy but does not inhibit p53/p73-regulated DRAM.
Activation of the PI3K/AKT signaling pathway inhibits the effect of DRAM-mediated autophagy on apoptosis in HCC cell lines. (a) An immunoblot assay was used to detect the activation of the PI3K/AKT pathway in 7702, HepG2, Hep3B and Huh7 cells starved (sta) for 48 h. (b) Cells were starved for 48 h with or without pre-treatment by transfection with PI3K siRNA (PI3K si). The ratio of apoptotic cells was determined by quantification of M30-positive cells. (c) An immunoblot assay was used to detect the effect of siRNA-induced PI3K knockdown on the expression of p53, p73, DRAM and LC3 I/II. (d) HepG2, Hep3B and Huh7 cells were transfected with DRAM siRNA (DRAM si) or co-transfected with DRAM and PI3K siRNAs; the cells were then starved for 48 h. The ratio of apoptotic cells was determined by quantification of M30-positive cells. (e) Cells were starved for 48 h with or without pre-transfection with PI3K si. The ratio of apoptotic cells was determined by quantification of M30-positive cells. The data are presented as the mean±S.D. of three independent experiments in (b), (d) and (e)
Starvation-induced DRAM cannot translocate to mitochondria to induce mitophagy in HCC cell lines
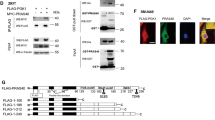
Our previous study demonstrated that mitochondrial DRAM can induce mitophagy, which then induces apoptosis.13 In this study, an immunoblot assay showed that starvation-induced DRAM and LC3 II accumulated in the extracted cytoplasm of 7702, HepG2, Hep3B and Huh7 cells; however, mitochondrial DRAM and LC3 II were detected in 7702 cells but not in the three HCC cell lines (Figures 7a and b). Moreover, we determined that both DRAM and GFP-LC3 puncta colocalized with HSP60 in 7702 cells in response to starvation using a confocal assay (Supplementary Figures 4 and 5). In the three HCC cell lines, neither DRAM nor GFP-LC3 puncta was colocalized with HSP60 in mitochondria (Supplementary Figures 4 and 5). These data suggest that starvation-induced DRAM could translocate to mitochondria and induce mitophagy in normal hepatocytes; however, in HCC cells, DRAM failed to induce mitophagy by localizing to mitochondria.
Phosphorylated AKT binds DRAM and prevents it from translocating to mitochondria in three HCC cell lines. (a) HepG2, Hep3B and Huh7 cells were starved (sta) for 48 h with or without pre-transfection with PI3K siRNA (PI3K si). The cytoplasm (C) and mitochondria (M) were extracted using Percoll gradient centrifugation. The levels of DRAM, LC3 I/II and p-AKT were evaluated by immunoblot assay. (b) 7702 cells were starved for 48 h, and an immunoblot assay was used to evaluate the levels of DRAM and LC3 I/II in the extracted cytoplasm and mitochondria. Anti-VDAC and anti-β-actin antibodies were used as controls for the extracted mitochondria and cytoplasm in (a) and (b). (c and d) p-AKT binds to DRAM in the cytoplasm but not in the mitochondria of HCC cells after starvation treatment for 48 h. HepG2, Hep3B (c) and Huh7 (d) cells were starved for 48 h, and the mitochondria and cytoplasm were extracted using Percoll gradient centrifugation. An anti-DRAM antibody was used to immunoprecipitate DRAM in the extracted mitochondria and cytoplasm. Total cell lysates were used as input or were immunoprecipitated with control IgG as indicated. DRAM and p-AKT were then analyzed by immunoblot assay using anti-DRAM and p-AKT antibodies. (e) Confocal microscopy was used to detect DRAM (green) and p-AKT (red) colocalization, as shown by the yellow sites in the cytoplasm of HepG2, Hep3B and Huh7 cells, in response to starvation for 48 h. DAPI was used to stain nuclei. Magnification, × 1000. (f) The diagram summarizes the interactions between DRAM-mediated mitophagy and the PI3K/AKT pathway in normal hepatocytes (blue) and HCC cells (red) (details are provided in the Results section)
Phosphorylated AKT inhibits the localization of DRAM to mitochondria, thereby inhibiting DRAM-mediated mitophagy in HCC cells
Here, using an anti-DRAM antibody to immunoprecipitate DRAM, we identified an interaction between p-AKT and DRAM in cytoplasm extracted from the three HCC cell lines following starvation (Figures 7c and d). An immunofluorescence assay also demonstrated that DRAM colocalized with p-AKT in the cytoplasm of the three HCC cell lines (Figure 7e). Moreover, we identified that PI3K knockdown using siRNA or LY294002 induced the appearance of DRAM and LC3 I/II in the mitochondria of the three HCC cell lines (Figure 7a and Supplementary Figure 6a). An immunofluorescence assay also revealed that inhibition of PI3K/AKT using LY294002 could induce the colocalization of either DRAM or GFP-LC3 puncta with HSP60, suggesting that activation of the PI3K/AKT pathway blocks the translocation of DRAM to mitochondria to induce mitophagy (Supplementary Figures 6b and c). As LY294002 treatment inhibited the PI3K/AKT pathway, we did not observe the colocalization of p-AKT and DRAM in the cytoplasm of the three HCC cell lines (data not shown). Taken together, our results suggest that in normal hepatoma cells, starvation-induced DRAM can induce apoptosis by translocating to mitochondria to induce mitophagy; however, in hepatoma cells starvation-induced p-AKT binds DRAM and sequesters it in the cytoplasm, thereby inhibiting the induction of apoptosis caused by DRAM-mediated mitophagy (Figure 7f).
Discussion
In this study, we determined that the effect of DRAM-mediated mitophagy on apoptosis is inhibited by activation of the PI3K/AKT signaling pathway in hepatoma cells in response to starvation. We believe that the finding that p-AKT binding to DRAM retards the translocation of DRAM to mitochondria is of considerable importance, as it links DRAM-mediated autophagic apoptosis to the PI3K/AKT pathway in hepatoma.
A clear relationship between the PI3K pathway and hepatoma has been found in many studies.23 Definitive evidence for the oncogenicity of PI3K was provided by the isolation of a constitutively active p110 isoform from the genome of the oncogenic avian retrovirus ASV16.24 PI3K can also be activated by several oncogenic growth factor receptors, such as platelet-derived growth factor and epidermal growth factor receptors, which highlights the participation of this pathway in the transduction of cancer-relevant cues.25, 26 As a key factor in the PI3K pathway, AKT is also linked to HCC. A recent study reported that the activation of AKT can predict poor prognosis in HCC.21
Our study further highlights the important role of AKT in hepatoma, as p-AKT inhibited the translocation of DRAM to mitochondria. Many previous studies have demonstrated that AKT can bind certain signaling proteins and translocate to many subcellular sites to regulate signaling pathways.27 In fact, we determined that starvation-induced p-AKT can translocate to mitochondria in HCC cells (Figure 7a). AKT can translocate from the cytosol to mitochondria, where it inhibits the opening of the permeability transition pore to maintain mitochondrial integrity. Thus, AKT has an anti-apoptotic role,28 as impaired mitochondrial integrity is an inducer of apoptosis.29 On the basis of our results, we hypothesize that both the p-AKT-induced prevention of DRAM translocation to mitochondria and the translocation of p-AKT to mitochondria partially contribute to the inhibitory effect of PI3K/AKT on apoptosis.
The effects of autophagy on hepatoma have been demonstrated in many studies. Mice with a heterozygous disruption of beclin-1 have a high frequency of spontaneous hepatoma.3 Expression of several autophagic genes (Atg5, Atg7 and Beclin-1) and their corresponding autophagic activity is decreased in HCC cell lines compared with that in normal hepatocytes.4 Moreover, therapeutic approaches aiming at increasing the autophagy level have been successfully tested in vitro and/or in mice.5, 6 Our previous study suggested that DRAM-mediated autophagy induces apoptosis in hepatoma cells.13 This study further revealed that activated PI3K/AKT can inhibit the induction of apoptosis by DRAM, suggesting that promoting DRAM-mediated autophagy together with the use of a PI3K/AKT inhibitor might be more effective for curing hepatoma. Moreover, this study also suggests that DRAM-mediated autophagy might be regulated by many signaling pathways, such as the PI3K/AKT pathway; however, further studies are needed to elucidate these complicated regulatory networks.
A previous study reported that DRAM can induce mitophagy based on detection of the mitophagy marker BNIP3.30 Later, we identified DRAM and LC3 I/II in extracted mitochondria, which suggests that DRAM translocates to mitochondria.13 A previous study used an immunofluorescence assay to detect mitophagy.31 In this study, we used the same method to determine that DRAM and GFP-LC3 puncta colocalize with the mitochondrial marker HSP60. Taken together, these results suggest that mitochondrial DRAM can regulate mitochondria function. In fact, Salem et al.30 have demonstrated that DRAM overexpression can affect the function of mitochondria. However, the means by which DRAM regulates mitophagy and mitochondrial function remains unclear. In addition to mitochondrial DRAM, DRAM also translocates to other organelles, including peroxisomes, lysosomes, the ER and autophagosomes.16 In these organelles, although the effects of DRAM on autophagy, apoptosis and cell survival or death are still unclear, DRAM can localize to multiple organelles, suggesting that the function of DRAM is rather complicated. Thus, additional studies are needed to elucidate the role of DRAM in cell fate in response to stimuli.
Overall, our results partially explain why promoting autophagy is effective for curing hepatoma. Our results also open new avenues for investigation into the mechanisms of autophagy-based therapy in hepatocellular cancer.
Materials and Methods
Cell culture and treatment
A human normal liver cell line (7702 (wild-type p53)) and human HCC cell lines (HepG2 (wild-type p53), Hep3B (p53 null) and Huh7 (p53A220G)) were grown in Dulbecco’s modified Eagle’s medium or in modified Eagle’s medium (Invitrogen, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum. The cells were seeded in 6- or 24-well plates and incubated in serum-deprived medium for 48 h. The cells were then transfected with plasmids encoding GFP-LC3 or siRNAs targeting DRAM and class I PI3K (the sequences of DRAM and class I PI3K siRNAs were obtained from references Crighton et al.14 and Nobukuni et al.32, respectively) using Fugene HD (Promega, Madison, WI, USA). BafA 1 (10 nM, Sigma Inc., St. Louis, MO, USA) and NH4Cl (10 mM, Sigma Inc.) were used to inhibit the autophagy flux. Cells were pre-treated with LY294002 (100 μM, Cell Signaling Inc., Beverly, MA, USA) for 2 h to inhibit PI3K activity; the medium was then replaced with normal culture medium for the following experiments.
Immunoblot assay
Cell lysates were subjected to western blot analysis as previously described.33 Briefly, total cellular lysates were separated on 10–15% SDS-PAGE gels, and the separated proteins were then transferred to a PVDF membrane. The protein blots were blocked with 5% milk and probed sequentially with specific primary antibodies and horseradish peroxidase-conjugated secondary antibodies. The detection of specific proteins on the blots was achieved with enhanced chemiluminescence (Pierce SuperSignal, Thermo Fisher Scientific Inc., Rockford, IL, USA), and images were captured on X-ray films. The primary antibodies were obtained from Abcam Inc. (Cambridge, MA, USA) (rabbit anti-DRAM polyclonal antibody (cat. no. ab68987)), Santa Cruz Inc. (Dallas, TX, USA) (goat anti-VDAC polyclonal antibody (N-18) and mouse anti-β-actin monoclonal antibody (C-2)), Cell Signaling Inc. (mouse anti-AKT monoclonal antibody (11E7), rabbit anti-p-AKT monoclonal antibody (D9E), rabbit anti-PI3K monoclonal antibody (C73F8) and rabbit anti-p-PI3K polyclonal antibody (cat. no. 4228)), Novus Biologicals Inc. (Littleton, CO, USA) (mouse anti-p62 monoclonal antibody (2C11)), Sigma Inc. (rabbit anti-LC3 polyclonal antibody (cat. no. L8918)) and Trevigen Inc USA. (Gaithersburg, MD, USA) (mouse anti-PARP monoclonal antibody (C2–10)).
Fluorescence microscopy and confocal assay
Frozen cells were fixed with 10% paraformaldehyde/PBS, incubated in 1% Triton X-100/PBS for 5 min, blocked with 3% BSA/PBS and probed with primary antibody. Alexa Fluor 488- or 594-conjugated secondary antibodies were used to amplify the signal. DAPI was used to stain nuclei. M30 immunoreactivity and calcein AM/PI were used to detect early and late apoptosis. Quantitative apoptosis analysis was performed by counting >1000 cells in each sample. The primary antibodies were obtained from Santa Cruz Inc. (mouse anti-DRAM monoclonal antibody (M3-P4B4)), Abcam Inc. (goat anti-HSP60 polyclonal antibody (cat. no. ab109873)) and Cell Signaling Inc. (rabbit anti-p-AKT monoclonal antibody (D9E)). The M30 mouse antibody was produced by our laboratory and was used to detect cleaved keratin 18 in early apoptotic cells. Confocal microscopy was used to detect mitochondrial DRAM and mitophagy based on the colocalization of DRAM or GFP-LC3 puncta with HSP60.31
Flow cytometry
The cells were fixed and stained with PI and analyzed on a Becton Dickinson FACScan (San Jose, CA, USA) as described previously.34 The percentage of cells with sub-G1 DNA content was used as a measure of apoptotic rate.
Real-time PCR
The RNeasy Mini Kit (Qiagen, Hilden, Germany) was used to isolate total RNA. Reverse transcription was performed using the SuperScript II First-Strand Synthesis System for RT-PCR (Invitrogen) to synthesize first-strand cDNA. SYBR green was used to detect dsDNA product during the real-time PCR reaction. The mRNA content was normalized to the housekeeping gene β-actin.13 The specific primer sequences used for real-time PCR were as follows: for DRAM, 5′-TCAAATATCACCATTGATTTCTGT-3′ (forward) and 5′-GCCACATACGGATGGTCATCTCTG-3′ (reverse) (the sequences of DRAM primers were obtained from reference Crighton et al.14); for β-actin, 5′-GCCCTGAGGCACTCTTCCA-3′ (forward) and 5′-CGGATGTCCACGTCACACTT-3′ (reverse).
Extraction of mitochondria and cytoplasm
The cytoplasm and mitochondria were isolated using Percoll gradient centrifugation based on previous methods.35 Briefly, the cells were Dounce homogenized on ice in M-SHE buffer (0.21 mol/l mannitol, 0.07 mol/l sucrose, 10 mmol/l HEPES-KOH (pH 7.4), 1 mmol/l EDTA, 1 mmol/l EGTA, 0.15 mmol/l spermine, 0.75 mmol/l spermidine and 1 mmol/l DTT) with freshly added protease inhibitors (1 μg/ml leupeptin, aprotinin, pepstatin A and 1 mmol/l phenylmethylsulfonyl fluoride). The nuclei were pelleted at 1200 × g, and the supernatant was centrifuged at 10 000 × g for 15 min at 4 °C to pellet mitochondria. After the 10 000 × g centrifugation step, the cytoplasmic fraction remained in the supernatant. The mitochondrial pellets were resuspended in 3% Ficoll 400/0.5 × M-SHE buffer, layered over 6% Ficoll 400/1 × M-SHE buffer and centrifuged at 10 400 × g for 25 min at 4 °C. The pellets were resuspended in M-SHE-0.3 mg/ml digitonin buffer at 4 °C for 15 min, centrifuged again at 10 500 × g for 15 min and subsequently washed once in M-SHE buffer. Extracts prepared by solubilizing mitochondria for 30 min on ice in lysis buffer (20 mmol/l HEPES (pH 7.4), 400 mmol/l KCL, 1 mmol/l EDTA, 5% glycerol, 0.5% Triton X-100, 2 mmol/l DTT and fresh protease inhibitors). Lysates were clarified by centrifugation at 16 000 × g at 4 °C for 1 h, concentrated to 10 mg/ml protein and stored at −80 °C or used immediately.
Immunoprecipitation assay
Cell lysates, extracted mitochondria and cytoplasm were pre-cleared with protein A/G PLUS-agarose beads (Santa Cruz Inc.) and were incubated with mouse anti-DRAM monoclonal antibody (M3-P4B4) (Santa Cruz Inc.) at 4 °C overnight. Immunocomplexes were separated by incubation with protein A/G agarose beads and were resolved by SDS-PAGE. Immunoblot assay was performed with rabbit anti-p-AKT monoclonal antibody (D9E) (Cell Signaling Inc.) and rabbit anti-DRAM polyclonal antibody (Abcam Inc., cat. no. ab68987).
Statistical analysis
All data are the result of at least three independent experiments and are expressed as the mean±S.D. The differences between groups were compared using Student’s t-test. Differences were considered significant at confidence levels of P<0.05, P<0.01 and P<0.001.
Abbreviations
- HCC:
-
hepatocellular carcinoma
- DRAM:
-
damage-regulated autophagy modulator
- LC3:
-
microtubule-associated protein light chain 3
- PI3K:
-
phosphatidylinositol 3-kinase
References
El-Serag HB, Rudolph KL . Hepatocellular carcinoma: epidemiology and molecular carcinogenesis. Gastroenterology 2007; 132: 2557–2576.
Parkin DM, Bray F, Ferlay J, Pisani P . Global cancer statistics, 2002. CA Cancer J Clin 2005; 55: 74–108.
Qu X, Yu J, Bhagat G, Furuya N, Hibshoosh H, Troxel A et al. Promotion of tumorigenesis by heterozygous disruption of the beclin 1 autophagy gene. J Clin Invest 2003; 112: 1809–1820.
Ding ZB, Shi YH, Zhou J, Qiu SJ, Xu Y, Dai Z et al. Association of autophagy defect with a malignant phenotype and poor prognosis of hepatocellular carcinoma. Cancer Res 2008; 68: 9167–9175.
Gao M, Yeh PY, Lu YS, Hsu CH, Chen KF, Lee WC et al. OSU-03012, a novel celecoxib derivative, induces reactive oxygen species-related autophagy in hepatocellular carcinoma. Cancer Res 2008; 68: 9348–9357.
Mazzanti R, Platini F, Bottini C, Fantappie O, Solazzo M, Tessitore L . Down-regulation of the HGF/MET autocrine loop induced by celecoxib and mediated by P-gp in MDR-positive human hepatocellular carcinoma cell line. Biochem Pharmacol 2009; 78: 21–32.
Mizushima N . Autophagy: process and function. Genes Dev 2007; 21: 2861–2873.
Mathew R, Karantza-Wadsworth V, White E . Role of autophagy in cancer. Nat Rev Cancer 2007; 7: 961–967.
Kroemer G, Jaattela M . Lysosomes and autophagy in cell death control. Nat Rev Cancer 2005; 5: 886–897.
Maiuri MC, Zalckvar E, Kimchi A, Kroemer G . Self-eating and self-killing: crosstalk between autophagy and apoptosis. Nat Rev Mol Cell Biol 2007; 8: 741–752.
Eisenberg-Lerner A, Bialik S, Simon HU, Kimchi A . Life and death partners: apoptosis, autophagy and the cross-talk between them. Cell Death Differ 2009; 16: 966–975.
Lockshin RA, Zakeri Z . Programmed cell death and apoptosis: origins of the theory. Nat Rev Mol Cell Biol 2001; 2: 545–550.
Liu K, Lou J, Wen T, Yin J, Xu B, Ding W et al. Depending on the stage of hepatosteatosis, p53 causes apoptosis primarily through either DRAM-induced autophagy or BAX. Liver Int 2013; 33: 1566–1574.
Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell 2006; 126: 121–134.
Crighton D, O'Prey J, Bell HS, Ryan KM . p73 regulates DRAM-independent autophagy that does not contribute to programmed cell death. Cell Death Differ 2007; 14: 1071–1079.
Mah LY, O'Prey J, Baudot AD, Hoekstra A, Ryan KM . DRAM-1 encodes multiple isoforms that regulate autophagy. Autophagy 2012; 8: 18–28.
Suzuki A, Hayashida M, Kawano H, Sugimoto K, Nakano T, Shiraki K . Hepatocyte growth factor promotes cell survival from fas-mediated cell death in hepatocellular carcinoma cells via Akt activation and Fas-death-inducing signaling complex suppression. Hepatology 2000; 32 (4 Pt 1): 796–802.
Nakanishi K, Sakamoto M, Yamasaki S, Todo S, Hirohashi S . Akt phosphorylation is a risk factor for early disease recurrence and poor prognosis in hepatocellular carcinoma. Cancer 2005; 103: 307–312.
Engelman JA, Luo J, Cantley LC . The evolution of phosphatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 2006; 7: 606–619.
Sarbassov DD, Guertin DA, Ali SM, Sabatini DM . Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005; 307: 1098–1101.
Schmitz KJ, Wohlschlaeger J, Lang H, Sotiropoulos GC, Malago M, Steveling K et al. Activation of the ERK and AKT signalling pathway predicts poor prognosis in hepatocellular carcinoma and ERK activation in cancer tissue is associated with hepatitis C virus infection. J Hepatol 2008; 48: 83–90.
Miyamoto S, Rubio M, Sussman MA . Nuclear and mitochondrial signalling Akts in cardiomyocytes. Cardiovasc Res 2009; 82: 272–285.
Chalhoub N, Baker SJ . PTEN and the PI3-kinase pathway in cancer. Annu Rev Pathol 2009; 4: 127–150.
Chang HW, Aoki M, Fruman D, Auger KR, Bellacosa A, Tsichlis PN et al. Transformation of chicken cells by the gene encoding the catalytic subunit of PI 3-kinase. Science 1997; 276: 1848–1850.
Escobedo JA, Navankasattusas S, Kavanaugh WM, Milfay D, Fried VA, Williams LT . cDNA cloning of a novel 85 kd protein that has SH2 domains and regulates binding of PI3-kinase to the PDGF beta-receptor. Cell 1991; 65: 75–82.
Hu P, Margolis B, Skolnik EY, Lammers R, Ullrich A, Schlessinger J . Interaction of phosphatidylinositol 3-kinase-associated p85 with epidermal growth factor and platelet-derived growth factor receptors. Mol Cell Biol 1992; 12: 981–990.
Fayard E, Xue G, Parcellier A, Bozulic L, Hemmings BA . Protein kinase B (PKB/Akt), a key mediator of the PI3K signaling pathway. Curr Top Microbiol Immunol 2010; 346: 31–56.
Miyamoto S, Murphy AN, Brown JH . Akt mediated mitochondrial protection in the heart: metabolic and survival pathways to the rescue. J Bioenerg Biomembr 2009; 41: 169–180.
Tan W . VDAC blockage by phosphorothioate oligonucleotides and its implication in apoptosis. Biochim Biophys Acta 2012; 1818: 1555–1561.
Salem AF, Whitaker-Menezes D, Lin Z, Martinez-Outschoorn UE, Tanowitz HB, Al-Zoubi MS et al. Two-compartment tumor metabolism: autophagy in the tumor microenvironment and oxidative mitochondrial metabolism (OXPHOS) in cancer cells. Cell Cycle 2012; 11: 2545–2556.
Tang Y, Chen Y, Jiang H, Nie D . Short-chain fatty acids induced autophagy serves as an adaptive strategy for retarding mitochondria-mediated apoptotic cell death. Cell Death Differ 2011; 18: 602–618.
Nobukuni T, Joaquin M, Roccio M, Dann SG, Kim SY, Gulati P et al. Amino acids mediate mTOR/raptor signaling through activation of class 3 phosphatidylinositol 3OH-kinase. Proc Natl Acad Sci USA 2005; 102: 14238–14243.
Li L, Jin R, Zhang X, Lv F, Liu L, Liu D et al. Oncogenic activation of glypican-3 by c-Myc in human hepatocellular carcinoma. Hepatology 2012; 56: 1380–1390.
Chen D, Padiernos E, Ding F, Lossos IS, Lopez CD . Apoptosis-stimulating protein of p53-2 (ASPP2/53BP2L) is an E2F target gene. Cell Death Differ 2005; 12: 358–368.
Chen D, Yu Z, Zhu Z, Lopez CD . The p53 pathway promotes efficient mitochondrial DNA base excision repair in colorectal cancer cells. Cancer Res 2006; 66: 3485–3494.
Acknowledgements
This work was supported in part by National Natural Science Foundation of China (81272266, 81361120401, 81100288 and 81071843).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by GM Fimia
Supplementary Information accompanies this paper on Cell Death and Disease website
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Liu, K., Shi, Y., Guo, X. et al. Phosphorylated AKT inhibits the apoptosis induced by DRAM-mediated mitophagy in hepatocellular carcinoma by preventing the translocation of DRAM to mitochondria. Cell Death Dis 5, e1078 (2014). https://doi.org/10.1038/cddis.2014.51
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.51
Keywords
This article is cited by
-
The role of the apoptosis-related protein BCL-B in the regulation of mitophagy in hepatic stellate cells during the regression of liver fibrosis
Experimental & Molecular Medicine (2019)
-
p53-Dependent PUMA to DRAM antagonistic interplay as a key molecular switch in cell-fate decision in normal/high glucose conditions
Journal of Experimental & Clinical Cancer Research (2017)
-
N-Myc and STAT Interactor regulates autophagy and chemosensitivity in breast cancer cells
Scientific Reports (2015)