Abstract
Chemoresistance hinders the curative cancer chemotherapy. To define the role of the DNA methylation-regulated microRNA (miR) genes in the chemoresistance of bladder cancer, we performed both DNA methylomic and miRomic analyses of a multi-chemosensitive (5637) versus a multi-chemoresistant (H-bc) cell line and found that miR-193a-3p is hypermethylated/silenced in 5637 and hypomethylated/expressed in H-bc cells. A forced reversal of its level turned around the chemoresistance in the cultured cells and the tumor xenografts in nude mice. Three of its targets: SRSF2, PLAU and HIC2, work in concert to relay the miR-193a-3p’s impact on the bladder cancer chemoresistance by modulating the activities of the following five signaling pathways: DNA damage, Notch, NF-κB, Myc/Max, and Oxidative Stress. In addition to the mechanistic insights in how the newly identified miR-193a-3p/SRSF2,PLAU,HIC2/five signaling pathway axis regulates the chemoresistance of bladder cancer cells, our study provides a new set of diagnostic targets for the guided personalized chemotherapy of bladder cancer.
Similar content being viewed by others
Main
Bladder cancer (BCa) is the second most common and the most life-threatening urogenital cancer in men.1, 2 In addition to the high recurring rate, BCa is highly refractory to drug therapy and regarded as one of the most costly and difficult type of cancer to be contained.3 Although an eradication of primary lesions by chemotherapy is achievable in about half of the BCa patients, the diseases often recur and become refractory to the second round of chemotherapy.4 Once the chemoresistance is established, the cancer cells are resistant to multiple chemo-therapeutics, irrespective of the nature of the previous drug exposure.5 As other hallmarks of cancer, the chemoresistance varies drastically among the patients suffered from any given type of cancer, making both the chemoresistance prediction and the rational personalized chemotherapy very difficult.
In addition to the extensive genetic abnormality, cancer cells suffer from a wide range of the epigenetic defects that are more tightly linked to the gene expression pattern and therefore the cancerous phenotypes. The following entities constitute the epigenetic interface of the human genome: DNA methylation, histone modification, noncoding RNA (including microRNAs (miRs)), the nucleosome organization, and high-order chromatin structure. The miRs that aberrantly expressed in cancer make a significant contribution to cancerous behaviors, including chemoresistance.6 Taking the miRs in the chemoresistance state of BCa as examples, the following miRs have been implicated: miR-34a,7 miR-21,8 miR-30d, miR-181, and miR-199a-5p.9 DNA methylation is the best-characterized epigenetic mechanism, underlying the faithful cross-cell-division transmission of the gene transcription memory.10 The hypermethylated state of the promoter and enhancer regions tightly correlates with the transcriptionally silenced state of both protein-coding and non-coding RNA genes. Therefore determining of the DNA methylation state of the promoter regions, rather than the level of the corresponding RNAs or proteins, in patient samples promises a better way for both early detection and rationale personalized therapy of cancer. Approximately, 10% of the miR genes are co-localized with the CpG-rich region (CpG islands) in genome, the expression of which is negatively regulated by DNA methylation. Hence we confined our attention to this subset of miRs for their roles in BCa’s chemoresistance. In this report, we began with a bi-omic (miRomic and methylomic) analysis of a multi-chemosensitive (5637) versus a multi-chemoresistant BCa cell line (H-bc) for the differentially expressed/methylated miR genes. The miR-193a-3p, one of the most differentially expressed/methylated CpG island containing miRs identified, was subjected to a systematic investigation for its role and the underlying mechanisms in the BCa’s multi-chemoresistance.
Result
Expression of the DNA methylation-regulated miR-193a gene positively correlates with the multi-chemoresistance of BCa cells
The drug dose for 50% cells killed by the following drugs: Pirarubicin (Pi), Paclitaxel (Pa), Adriamycin (Ad), and Epirubicin Hydrochloride (EH) after a treatment of 72 h, was determined in the following five BCa cell lines: 5637, T24, EJ, H-bc, and Biu87 (Figures 1a and b). Judged by the fold difference over the lowest IC50, 5637 was the most multi-chemosensitive cell line, with the lowest IC50 to four of five drugs, while H-bc was the most resistant cell line with its relative IC50 by 8.32 to 36.96 folds higher than 5637 cells (Figure 1c). From a RNA-seq based miR-omic analysis of 5637 and H-bc cell lines (Figure 2b and data not shown), 83 miRs were found differentially expressed by no less than two folds: 37 higher and 45 lower in 5637 than H-bc cells, respectively. Among the 20 miR genes that are co-localized with the CpG island, the miR-193a-3p differentially expressed between 5637 and H-bc cells at the top range in both RNA-seq omic (Figure 2b) and a qRT-PCR analysis (Figures 2c and d). The hypermethylated state in 5637 and hypomethylated state in H-bc cells of the miR-193a gene suggested by the methyl-capture seq analysis11 (Figure 2e, omic data not shown) were confirmed by a bisulfite conversion sequencing (BSP) analysis (Figures 2f and g): It is hypermethylated in 5637 (the average CpG methylation: 78.6%) and barely methylated in H-bc (the average CpG methylation: 4.3%). In conclusion, the differential state of the miR-193a gene at both DNA methylation and expression levels tightly correlates with the multi-chemoresistance of BCa cells.
Chemoresistance profiling of five BCa cell lines. (a) The experimental scheme. (b) The IC50 of four indicated chemotherapeutics of BCa cells. The percentage of the relative cell survival rates over the mock treatment was calculated and plotted against l g ng/ml of drug. (c) The relative IC50 (-fold) with the lowest IC50 (5637 cell line) are presented in Table
Differential methylation/expression of the miR-193a gene in 5637 versus H-bc cells. (a) The experimental scheme. (b) The top differentially expressed CGI containing miRs from the seq-based miRomic analysis in Heatmap. The relative expression (fold) of both miR-193a-3p and -5p between 5637 and H-bc, validated by qRT-PCR analysis (c) and summarized in the Table (d). (e) DNA methylation state of the miR-193a gene from the Methyl-cap-seq analysis, which are indicated as dark peaks. CpG dinucleotides and the BSP primers are also shown. (f) The original sequencing result of both strands of the PCR product of the bisulfite-converted DNA are shown. The percentage of the CpG methylation is summarized in the table (g) and in the plot (h)
SRSF2, PLAU and HIC2 are direct targets of miR-193a-3p in BCa cells
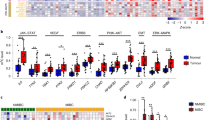
The number of the genes potentially regulated by one miR ranges from several hundreds to a couple of thousands. Among the predicted target genes of miR-193a-3p that were picked up by no less than three out of four commonly used predicting methods: miRDB (166 entries identified), miR base (380 entries), targets can (443 entries), target miner (243 entries), HIC2 (hypermethylated in cancer 2) gene, along with two known miR-193a-3p’s targets: SRSF2 (serine/arginine-rich splicing factor 2)12 and PLAU (plasminogen activator, urokinase),13, 14, 15 were found to be expressed in an opposite manner of the miR-193a-3p at both RNA (RNA-seq based omic: Figure 3b and qRT-PCR analysis, Figure 3c) and protein levels (Figure 3d). Furthermore, a miR-193a-3p mimic transfection brought down the levels of all three genes in 5637 cells, and their levels were raised in the antagomiR-transfected H-bc cells (Figures 3e and f).
Expression of the miR-193a-3p’s target genes in a reverse pattern of miR-193a-3p. (a) The experimental scheme. Expression of SRSF2, HIC2 and PLAU genes in 5637 versus H-bc cells at the mRNA level from the RNA-seq analysis (b) and by qRT-PCR (c) as well as at the protein level (d). Expression of SRSF2, HIC2 and PLAU genes in the miR-193a-3p mimic (3PM)-transfected 5637 and miR-193a-3p antagomiR (3PA)-transfected H-bc cells at the protein level by western blotting analysis (e) and mRNA level by qRT-PCR (f). (g) The schematic map of the pGL3-based luciferase reporter constructs where the UTR region (3′-UTR) of HIC2 or PLAU gene was put at the downstream flank of luciferase gene. The luciferase activity (fold) of the pGL3 with PLAU-UTR sequence (h) or HIC2 (i) relative to VEC (with no UTR seq) was determined in the cells transfected with the miR-193a-3p mimic (3PM), antagomiR(3PA) or Scramble (NC). The representative results from three independent experiments are shown. Error bars represent S.E.M. **P<0.01; by Student’s t-test
For the proof that HIC2 and PLAU genes are direct targets of miR-193a-3p, both 3′-UTR regions were put at the downstream of the firefly luciferase gene in pGL3 (Promega, Madison, WI, USA) to create pGL3-HIC2 UTR and pGL3-PLAU UTR constructs (Figure 3g). Both constructs and pGL3 were transfected into 5637 and H-bc cells, respectively, for the functional state of miR-193a-3p in cells. Both UTR-contained constructs gave a significantly higher luciferase activity in 5637 than H-bc cells in an opposite pattern of miR-193a-3p’s expression, in contrast to the no difference state of the pGL-3-resulted luciferase activities in both cell lines (Figures 3h and i). Moreover, the miR-193a-3p mimic transfection repressed the luciferase activities of both 3′-UTR containing but not pGL-3 constructs in 5637 cells, and the reverse was found in the antagomiR-transfected H-bc cells (Figures 3h and i). In conclusion, similar to SRSF2,12 both PLAU and HIC2 are the true direct targets of miR-193a-3p in BCa cells. The miR-193a-3p’s effect on the BCa chemoresistance is likely to be realized by its repression of these three genes.
miR-193a-3p rather than -5p dictates the multi-chemoresistance of BCa cells
We have previously reported the positive involvement of miR-193a-3p via repression of SRSF2 expression in the 5-FU chemoresistance of hepatocellular carcinoma (HCC).12 The miR-193a-5p’s regulatory role in both cell survival and chemosensitivity was also reported, probably via repression of TP73 expression.16 Both SRSF2 and TP73 proteins were indeed significantly lower in H-bc than in 5637 cells (not shown) and were upregulated by the mimic transfection in 5637 and downregulated by the antagomiR transfection in H-bc cells (Figures 4c and e), well matching the level (functional state too) of miR-193a-3p and -5p (Figure 1). A miR-193a-3p but not -5p mimic transfection are capable to reduce the drug-triggered cell death in 5637 cells by 0.2–1.3-fold in a drug-specific manner (Figure 4b). The same conclusion was drawn from the analysis in the antagomiR-transfected H-bc cells (Figure 4d). Therefore, miR-193a-3p but not -5p contributes to the BCa’s multi-chemoresistance, although both express in a same pattern.
The effect of both miR-193a and its target genes on the BCa chemoresistance. (a) The experimental scheme. (b) The relative cell survival by the IC50-dosed indicated drugs in the miR-193a-3p (3PM) or miR-193a-5p (5PM) mimic versus the mock (NC) of 5637 cells in plot. (c) The relative level of both SRSF2 (-3p target) and TP73 (-5p target) normalized with the GAPDH level, in the same set of 5637 cells in panel (b). (d) The relative cell survival by the IC50-dosed indicated drugs in miR-193a-3p (3PA) or miR-193a-5p (5PA) antagomiR versus the mock (NC) in H-bc cells in plot. (e), The relative level of SRSF2 (-3p target) and TP73 (-5p target) proteins normalized with the GAPDH level, in the same set of H-bc cells in panel (d). (f) The relative cell survival of the siRNA or 3PM-transfected or NC-transfected 5637 cells under the IC50-dosed Pa and Ad, respectively. (g) The protein level of the downstream genes in the same set of cells in panel (f), by western blotting analysis. (h) The relative cell survival of 5637 cells transfected with the indicated transfection under the IC50-dosed Pa and Ad, respectively. (i) The protein level of the experiment in panel (h) (Pa-treated group), by western blotting analysis. (j) The summary of the effects on the cell survival of the IC50-dosed drug-trigger cell death by miR-193a-3p’s target genes. (k) The relative cell survival under the treatment of the IC50-dosed Pa and Ad in H-bc cells transfected with the SRSF2-GFP, PLAU-GFP expression constructs, 3PA or NC, respectively. (*P<0.05; **P<0.01)
We then individually repressed each miR-193a-3p’s target genes by a siRNA transfection (Figure 4g) in 5637 cells, followed by the measurement of the Pa- or Ad-triggered cell death. With the miR-193a-3p mimic transfection as a reference, a compatible impact was observed. Repression of either HIC2 or PLAU level gave a stronger effect than the repression of SRSR2 gene (Figure 4f). The siRNA-mediated co-repression of both SRSF2 and PLAU or all three genes caused a further reduction of Pa-triggered but not Ad-triggered cell death (Figure 4l). Intriguingly, only the co-repression of both SRSF2 and HIC2 led to a further reduction of the Ad-triggered (but not Pa-triggered) cell death in 5637 cells (Figures 4h and k). Therefore, the synergistic effect of miR-193a-3p’s target genes on the BCa’s chemoresistance was in both gene-specific and drug-type-specific fashion. Conversely, a forced upregulation of both SRSF2 and PLAU proteins by transfection of the constructs where the GFP-tagged genes are under the potent mammalian promoter sensitized H-bc cells to the Ad-triggered cell death, but neither mock nor the GFP vector transfection did. Compatible to the 3PA’s effect, the forced upregulation of both genes had no effect on the Pa-triggered cell death (Figure 4i). An expected expression of the exogenously introduced SRSF2-GFP and HIC2-GFP proteins in H-bc cells was confirmed by examination under the fluorescent microscope (Supplementary Figure S1B). In conclusion, the miR-193a-3p’s effect on the BCa chemoresistance is indeed significantly accomplished by repression of these three genes, the subset of genes involved contributing to the drug-specific nature of the BCa chemoresistance.
miR-193a-3p regulates the activities of the chemoresistance-associated signaling pathways in the content of the BCa multi-chemoresistance
For further mechanistic insights into the BCa chemoresistance, we determined in both 5637 and H-bc cells the activities of the following 16 signaling pathways using the Qiagen’s pathway reporter systems: Oxidative Stress, DNA Damage, NF-κB, Hypoxia, ER Stress, Heavy Metal Stress, Heat Shock, Glucocorticoid, JNK, Xenobiotic, Wnt, Notch, TGF-β, Cell Cycle/pRb-E2F, Myc/Max, and MAPK/ERK (Figure 5a). Activities of seven pathways differed by more than two-folds between 5637 and H-bc cells, which might have a significant role in BCa chemoresistance. The pathways with higher activities in 5637 than in H-bc cells were DNA damage, NF-κB, Myc/Max and Heavy/Metal Stress pathways. The former three pathways were downregulated in the mimic-transfected 5637 cells and upregulated in the antagomiR-transfected H-bc cells (Figure 5b and Supplementary Figure S2). A reverse effect was observed on Oxidative Stress, Notch and Hypoxia pathways. The former two were activated in the mimic-transfected 5637 and repressed in the antagomiR-transfected H-bc cells (Figure 5c and Supplementary Figures S3A–F). We then individually repressed SRSF2, PLAU and HIC2 in 5637 cells by siRNA transfection for the impact on pathway activities. Only the activities of both Notch and Oxidative Stress were elevated in the SRSF2 siRNA-transfected 5637 cells. The pathways regulated by PLAU were DNA Damage, NF-κB and Myc/Max in a positive fashion. The pathways regulated by HIC2 were Myc/Max in a positive fashion and Oxidative Stress/Notch signaling in a negative fashion (Figure 5d and Supplementary Figures S2G–I). Compatible to the effect on the drug-triggered cell death, the influence of the miR-193a-3p’s target genes on the pathways are in a gene-specific manner.
The signaling pathways regulated by miR-193a and its downstream genes. (a) The experimental scheme. (b) The relative activities (mean±S.D.) of seven pathways that differed by more than two-fold between 5637 and H-bc cells. (c) The relative activities of these seven pathways indicated in panel (b). The grey cells indicate the pathways that failed to respond in an expected manner at least in one case. (d) The pathways regulated by HIC2, PLAU and SRSF2 in 5637 cells. The grey cells indicate the pathways that failed to respond in an expected manner. (e) The STRING network models drawn with cytoscape 2.8.3. Left: Initial network of eight seeds (three miR-193a-3p targets and five master transcription factors for each affected pathway generated with the STRING one-step direct path algorithm. Right: expanded connected network with CCNT1, HDAC9, HDAC6 and PCAF as hubs. Edge: the arrow-headed line indicates positive regulation via binding, T-headed line indicates repression via binding. Solid lines indicate the interaction with high confident experimental evidence support. Dashed lines indicate the potential interactions. The green circles indicate the upregulated genes and the red indicate downregulated genes in 5637 relative to H-bc cells (Based on RNA-seq data). (Notch, Myc, DNA damage (DD), NF-κB and Oxidative Stress (OS))
For the further mechanistic understanding, we searched for the interactions among the three miR-193a-3p’s targets (PLAU, SRSF2 and HIC2 genes) and the master transcription factor genes for these five signaling pathways (Figure 5d): NRF1 for Oxidative stress, MYC for Myc/Max, RBPJ for Notch, TP53 for DNA Damage and NFKB1 for NF-κB pathways from the STRING database of the known protein–protein interactions (PPI) interactions.17 Using one-step direct path algorithm, a direct link between PLAU and TP53 (DNA-damage response pathway) or NFKB1 (NF-κB pathway) was shown. A direct physical interaction between TP53 and MYC protein (Myc/Max pathway), suggested by a proteomic analysis, may contribute to the PLAU’s effect on the Myc/Max pathway (Figure 5e). Via both literature mining and the expression analysis of these eight genes in the RNA-seq data sets of 5637 and H-bc cells (not shown), we built a more sophisticated connection network. Three hub genes connecting both SRSF2 and PLAU to NRF1, RBPJ and TP53 genes, respectively, were all involved in the regulation of the acetylation state of both histones and non-histone proteins: PCAF (histone acetyltransferase b) and two histone deacetylases (HDAC6 and HDAC9). The hub gene connecting HIC2 protein to MYC (Myc/Max) pathway is CCNT1, encoding a CDK9-associated C-Type protein with a positive regulatory role in transcript elongation.18 The recent finding that HIC2 forms a complex with SRSF2 via both CCNT1 and Cdk919, 20, 21 may explain why both HIC2 and SRSF2 genes share the repressing role on both Notch and Oxidative pathways.
miR-193a-3p promotes both growth and Pa chemoresistance of the 5637- and H-bc-derived tumor xenografts in nude mice
To minimize the inter-mouse bias, 5637 (1.5 × 107 cells/site) or H-bc (0.7 × 107 cells/site) cells were subcutaneously injected at the four back sites of each six mice each. An intratumor injection of miR-193a-3p/-5p agomiR/antagomiR or the scramble sequence control (Mock) or phosphate-buffered saline (PBS) into the 5637/H-bc-derived tumors was initiated on the fourth day and repeated four times once in 2 days. The intraperitoneal injection of PBS or Pa was started on day 6 into three mice each in either the 5637 or H-bc group and repeated four times once in 2 days (Figure 6a). The tumor mass was weighed on day 25 at the end of this study. With half less cells injected, the H-bc-derived tumors were significantly heavier than 5637-derived tumors (0.81 g/0.53 g=1.53 in tumor weight) (Figures 6c and d), suggesting a miR-193a’s promoting role for the in vivo in tumor growth. This conclusion was supported by the experiments where both 5637 and H-bc tumor xenografts were established in the same mice (not show). An intratumor injection of miR-193a-3p but not -5p agomiR in comparison with the mock into the 5637 cell-derived tumor xenograft resulted in a bigger tumor mass of 5637 cell-derived tumor xenografts (0.81/0.53=1.53). The reverse observations were made from the antagomiR in H-bc tumor mice (0.13/0.40=0.33 in tumor weight). Therefore, the miR-193a-3p but not -5p is capable to promote the in vivo tumor growth. Consistent with the observation that H-bc was more Pa-resistant than 5637 cells in cultured cells (Figure 1), an intraperitoneal injection of Pa caused the 5637 tumors in a much smaller size than the H-bc tumor: the tumor weight ratio of the Pa treated over PBS treated is 0.283 (5637) versus 0.40 (H-bc) (Figure 6d). To separate the tumor growth-promoting effects from the Pa resistance-enhancing effects, the tumor weights of the Pa group where miR-193a-3p, -5p and mock-transfected tumor xenografts were established in the same individual mice were compared. The ratios of tumor weight of the miR-193a-3p agomiR/Mock in 5637 mice (3.42) were greater than that in the -5p agomiR/Mock counterpart (1.57), indicating that miR-193a-3p compromised the Pa’s tumor-inhibition capability. The same was concluded from the experiments with the antagomiR injection into the H-bc-derived tumor xenografts: the relative tumor weight of the -3p antagomiR/Mock (0.33) was smaller than that of the miR-193a-5p antagomiR/Mock: (1.1, Figures 6c and d). Again, neither miR-193a-5p agomiR nor antagomiR altered the growth of the Pa-treated 5637 or H-bc tumors.
The effect of miR-193a-3p and miR-193a-5p on both tumor growth and PA chemoresistance of 5637- and H-bc-derived xenografts in nude mice. (a) The experimental scheme, 5637 cells or H-bc cells were subcutaneously injected at four points at the back of each nude mice, respectively. From day 4, PBS, the indicted agomiR/antagomiR or the scramble sequence control (Mock) was intratumorly injected into 5637/H-bc, respectively, once every 2 days for five times as indicated. From day 6, each of the three mice for 5637 or H-bc received the intraperitoneal injection of PBS or Pa once every 2 days for five times, respectively. (b) The image of the representative mice with tumors on the day 25. (c) The mean±S.D. of tumor weight of the tumor from same treatment was calculated and plotted, *P<0.05 and summarized in the table. (d and e) The 5637 or H-bc tumor tissues from each group were fixed on one same slide and immunostained with the indicated antibodies. Levels of Ki67, SRSF2 and TP73 proteins in each indicated tumor tissues were scored and summarized
Further confirmation of the miR-193a’s role in Pa resistance of BCa came from the immuno-histological analysis of SRSF2 (a miR-193a-3p’s target), TP73 (a -5p’s target) and Ki67 (an indicator for cell proliferation) in the tumor sessions of the Pa-treated versus PBS-treated mice (Figure 6e and Supplementary Figure S4). The intratumor injection of either miR-193a-3p or -5p’s agomiR (into 5637)/antagomiR (into H-bc tumor) indeed led the expected changes of both SRSF2 or TP73 levels in tumor slides (Figure 6e), which consolidate the conclusion that miR-193a-3p but not miR-193a-5p has a profound positive effect on both the growth and chemoresistance of the BCa cell-derived tumor xenografts in nude mice.
Discussion
Aberrant miR-193a expression has been reported in all the types of cancer examined, including colorectal cancer,22 non-small cell lung cancer (NSCLC),23 myeloid leukemia24 and Wilms’ tumor blastema.25 The transcription factors implicated in the regulation of miR-193a transcription include XB130,26 p63,16 Max and RXR.13 As a typical CpG island-containing gene, its transcription is also under a regulation of the DNA methylation state of the promoter region and activated by a 5’ aza-2’-deoxycytocine-mediated demethylation.12 The tumor-suppressor role of miR-193a-3p has been reported in NSCLC27 and epithelial ovarian cancer cells.28 On the other hand, miR-193a-3p can also promote both in vivo growth and chemoresistance of HCC12 and of BCa in the present study. The functional disparity in cancer biology of miR-193a-3p in literature is likely attributed to the system difference of studies concerning the type of cancer, the experimental designs and whether the in vivo studies involved. It is also very likely that miR-193a-3p can act as both a tumor suppressor and oncogene in a biological and pathological content-dependent manner.
A miR executes its biological function via repression in a sequence-specific manner of up to a couple of thousands protein-coding genes at both stability and translation levels of mRNAs. In this study, we have defined the roles of PLAU, HIC2 and SRSF2 genes, three direct targets of miR-193a-3p, that work in concert to relay the miR-193a-3p’s impact on the BCa multi-chemoresistance (Figure 4). PLAU encodes the urokinase-type plasminogen-activator protein and actively involves in cell migration, invasion and angiogenesis processes. Its level is much higher in an Ad-resistant than its sensitive counterpart in breast cancer.29 There was a report that the siRNA-mediated repression of PLAU sensitizes HEK-293 cells to the UV-, cisplatin- and detachment-triggered apoptosis through DNA-damage pathway.30, 31 However, the opposite was found in our study, that is, a siRNA-mediated repression of PLAU enhanced rather than repressed the multi-chemoresistance of 5637 cells (Figure 4). HIC2 encodes a putative transcriptional factor, the genetic aberration of which is associated with both digeorge syndrome,32 and systemic lupus erythematosus33 in humans. However, its role in chemoresistance of cancer is, for the first time, described here. SRSF2 protein is a member of the serine/arginine (SR)-rich family of pre-mRNA splicing factors, a critical component of the spliceosome executing both splicing and nuclear-cytoplasmic transportation of mature mRNA.34 Its role in the miR-193a-3p’s effect on the 5-FU chemoresistance in HCC12 is essentially confirmed in the BCa multi-chemoresistance in the present study. We also demonstrated a gene-specific as well as a drug-type specific nature of the additive (or synergistic) effect of these three genes on the BCa chemoresistance (Figure 4h).
Although the signaling pathways affected by the target therapeutics are well established, the pathways challenged by the conventional chemotherapeutics remain undefined. Pa, a cytoskeleton drug is expected to disturb different pathways from Ad, a genotoxic drug. The assumption that a key set of pathways involved in the BCa chemoresistance have significantly different activities between the multi-chemosensitive 5637 and the multi-chemoresistant H-bc cells was confirmed; 7 of the 16 signaling pathways tested displayed different activities by more than two-fold in this pair of BCa cells (Figure 5b), 5 of which were regulated by miR-193a-3p (Figure 5c) and its target genes (Figure 5d) in an expected fashion. By a network analysis using three miR-193a-3p’s target genes and five master transcription factors as cues, a number of significant interactions among these eight genes have been found (Figure 5e). HIC2’s capability to form a complex with SRSF2 via a physical interaction with both CCNT1 and Cdk919, 20, 21 may explain why these two miR-193a-3p’s target genes repress both Notch and Oxidative pathways in a similar manner. In addition, HDAC6 deacetylates, stabilizes SRSF2 protein and therefore enhances its function.35 Three hub genes connecting SRSF2 to the master transcription factors RBPJ (Notch) and NRF1 (Oxidative stress) are two histone deacetylase (HDAC6 and 9) and one histone acetyl-transferase (PCAF). These three proteins work in concert to delicately control the acetylation state of both core histones and non-histone proteins.36 PCAF can also interact with RBPJ37 or NRF138 for an activation of both pathways. HIC2, but not SRSF2, acts as a positive regulator of MYC/Max pathway, probably via HIC2’s interaction with CCNT1 and in turn activates the positive transcription elongation factor b (P-TEFb)21.39 The transcription of the miR-193a gene is regulated by Max and RXR.13 The regulatory role of both HIC2 and PLAU on MYC/max pathway in the context of the multi-chemoresistance of BCa cells (Figures 4 and 5) suggests a negative regulating loop between the miR-193a-3p and MYC/Max pathway.
As summarized in Figure 7, we have identified a miR-193a-3p centered axis that dictates the BCa’s multi-chemoresistance, expression of which was under the negative control of the DNA methylation. Via its repressive effect on three target genes (and others) and in turn the five signaling pathways, the miR-193a-3p promotes the multi-chemoresistance (Pa, Pi, EH and Ad studied in this report) and the in vivo tumor growth of BCa cells. For further mechanistic insights, we have informatically identified the key links that connect miR-193a-3p via three of its target genes (SRSF2, PLAU and HIC2) to the five signaling pathways and therefore the chemoresistant phenotype of BCa cells (Figures 5e and 7b). Our study provides a new set of diagnostic targets for the guided personalized chemotherapy of BCa.
The working model for the miR-193a’s role in regulation of the chemoresistance of BCa by repression of HIC2, PLAU and SRSF2. (a) Expression of SRSF2, HIC2 and PLAU genes repressed by DNA methylation-regulated miR-193a-3p, is positively correlate with the chemoresistance to four indicated chemotherapeutics. (b) A network model. Lines signify the potential interactions between genes obtained from the STRING database. The green nodes indicate the upregulated genes and the red nodes indicate the downregulated genes in 5637 relative to H-bc cells. (Notch, Myc, DNA damage (DD), NF-κB and Oxidative Stress (OS)). (c) Relative expression level from RNA-seq data of the genes are summarized in the table
Materials and methods
Studies in cell culture
Cell lines: Five transitional carcinoma cell lines of BCa used in this study: EJ (established by Marshall CJ in 197740), T24 (ATCC NO. HTB-4), 5637 (ATCC NO. HTB-9), H-bc (established by cancer research Institute of Kunming Medical College, 1986), and Biu87 (established by department of Urology of Beijing Medical University in 198741) were purchased from the Chinese Academy of Sciences Committee on Culture Collection Cell Bank, Shanghai Institutes for Biological Sciences, Shanghai, China. Cells are cultured in RPMI1640 (Invitrogen, Carlsbad, CA, USA)+10% fetal bovine serum (Invitrogen) and 1% glutamine at 37 °C in 5% CO2.
The mimic/antagomiR/siRNA/overexpression plasmids transfection: all the mimic, antagomiR, siRNA, and the scramble sequence control (NC) as well as riboFECT CP transfection kit were supplied by Guangzhou Ribobio (Guangzhou, China). The mammalian expression constructs for PLAU (EX-F0073-M98-5) and SRSF2 (EX-F0622-M98-5) with GFP tag were supplied by Guangzhou Fulengen (Guangzhou, China). Transfection of both ribonucleic acid reagents or plasmids mentioned above and the reporter plasmids in a Cignal Finder Pathway Reporter package (Qiagen, Hilden, Germany) was performed according to the manufacturer’s instruction.
The luciferase reporter assay
A full length of the human PLAU 3′-untranslated region (932 bp) and partial length of HIC2 3′-UTR (1131 bp, 1862-2992 from a 4719-bp full length) with the miR-193a-3p targeting sequence were cloned at the downstream of the firefly luciferase gene in pGL3 (Invitrogen) to construct pGL3-luc-PLAU and pGL3-luc-HIC2, respectively. All the constructs were confirmed by restriction digestion.
Cells were seeded into 96-well plates at around 1 × 104 cells per well and transfected with a mixture of 50 ng pGL3-luc-PLAU or pGL3-luc-HIC2, 5 ng Renilla plus 5 pmol mimic or scramble control (NC) nucleotides, with the riboFECT CP transfection reagents according to the manufacturer’s instruction. Both firefly and Renilla luciferase activities were measured 18 h after transfection by the Dual-Luciferase Reporter Assay System (Promega) using a Promega GloMax 20/20 luminometer. The relative firefly luciferase activities were normalized with the Renilla luciferase activities as a for transfection efficiency.
The pathway luciferase reporter constructs (Supplementary Figure S2A): (1) the negative control construct: the firefly luciferase gene is under the control of the minimal promoter. (2) the pathway reporter construct: a tandem repeat of the cognate consensus motif that is recognized by each master transcription factor for the corresponding pathway was placed at the upstream of the minimal promoter in the construct 1. (3) the positive control construct: the firefly luciferase gene is under the control of CMV promoter and (4) the internal control construct. The firefly luciferase gene in construct 3 was replaced with the Renilla luciferase gene. The analysis was carried out according to the manufacturer’s instruction (Qiagen).42 Briefly, the cells were transfected in triplet with each firefly luciferase reporter construct in combination with the Renilla luciferase construct using ribo FECT CP transfection reagent, and both luciferase activities in cell extracts at 24 h after transfection were measured by a Promega Dual-Luciferase Reporter assay (Promega) using a Promega GloMax 20/20 luminometer.43 Firefly luciferase activities from each set were normalized to the activity of Renilla luciferase to control the inter-transfection bias. The relative luciferase activities (luciferase unit) of the pathway reporter over the negative control in the transfected cells were calculated as a measurement of the pathway activity.
Chemotherapeutics: All the chemotherapeutic drugs used are of the clinic grade44, 45 (NCI Dictionary of Cancer Terms, http://www.cancer.gov/dictionary), Pi: Pirarubicin hydrochloride (Wanle, Shenzhen, China); Pa: Paclitaxel (Shuanglu, Beijing, China); Ad: Adriamycin (Haizheng, Zhejiang, China); EH: Epirubicin hydrochloride (Haizheng, Zhejiang, China).
Chemoresistance profiling (IC50 measurements): Cells in the logarithmic phase of growth were seeded in triplicate in 96-well plates at the density of 0.5 × 104/well and treated with four-fold serially diluted drugs for 72 h. Cell survival was then measured by a thiazolyl blue tetrazolium bromide (MTT, 490 nm reading)-based cell proliferation assay.46 Both the linear regression parameters and the IC50 (the concentration of drug required for 50% of cells to be killed) with the no-drug control as the reference were calculated. The relative chemoresistance was presented as the fold for each of the cell line over the lowest IC50.
RNA analysis
Total RNA was isolated using the TRIzol reagent (Tiangen Biotech Co., Ltd., Beijing, China). For mRNA analysis, the cDNA was made from total RNA by oligo-dT priming with a primeScript RT reagent kit (Tiangen Biotech Co., Ltd.) and the mRNA level of the following three genes (SRSF2, HIC2 and PLAU) was measured by qRT-PCR with gene-specific fluorescent Taqman probe together with the β-actin using a different fluorescence-labeled probe (provided by ShingGene, Shanghai, China) in the FTC-3000P (FUNGLYN BIOTECH INC, Toronto, ON, Canada). For miR analysis, the cDNA was synthesized with the specific stem-loop primer and quantified by SYBR Green-based real-time PCR assay. Using the 2−ΔΔCt method, the normalization with the U6 reads for miR or with β-actin for mRNA was performed before each’s relative level between 5637 and H-bc was calculated.47 The primers and probes for complementary DNA synthesis and qRT-PCR analysis are:
hSRSF2F: 5′-CAAGTCCAAGTCCTCGTCGG-3′
hSRSF2R: 5′-TGGATTCCCTCTTGGACACTG-3′
hSRSF2 probe: 5′ROX-CTCGTTCGCGGTCCAGGTCCC-3′ BHQ2 for SRSF2,
hHIC2F: 5′-GGTGACCAGGCATTGATGC-3′
hHIC2R:5′-GAGCTGCTTCGAGTGGCTG-3′
hHIC2 probe: 5′FAM- ACGCGCACCACCGGAGTGC-3′ BHQ1 for HIC2
hPLAUF: 5′-CCATCTACAGGAGGCACCG-3′
hPLAUR: 5′-GCGACCCAGGTAGACGATG-3′
hPLAU probe: 5’ROX-CTCATCAGCCCTTGCTGGGTGATC-3’BHQ2 for PLAU
hACTBF: 5′-GCCCATCTACGAGGGGTATG-3′
hACTBR: 5′-GAGGTAGTCAGTCAGGTCCCG-3′
hACTB probe: 5′HEX-CCCCCATGCCATCCTGCGTC-3′ BHQ1 for β-actin.
Western blotting analysis
Cell lysates with 1 × SDS loading buffer (60 mM Tris-HCl, pH6.8, 2% SDS, 20% glycerol, 0.25% bromphenol blue, 1.25%2-mercaptoethanol) were incubated at 100 °C for 10 min to facilitate the sample loading for the conventional western blotting analysis. The anti-SRSF2 (AP2800a), anti-HIC2 (AP18558c), anti-PLAU (AP8161b) and anti-GAPDH (AM1020a) were provided by Wuxi phama, Shanghai, China. The target proteins were then probed with anti-rabbit IgG peroxidase-conjugated antibody (LP1001b), or HRP goat anti-mouse IgG antibody (LP1002a) (All antibodies are from Abgent, San Diego, CA, USA) followed by an enhanced chemiluminescence reaction (Thermo Fisher Scientific, Waltham, MA, USA). The relative levels of proteins were quantified using densitometry with the Gel-Pro Analyzer (Media Cybernetics, Rockville, MD, USA). The target bands over the GAPDH band were densitometrically quantified and indicated under each band.
BSP analysis
Genomic DNA was isolated by a standard phenol/chloroform purification method, qualified by electrophoresis on an agarose gel, treated by ammonium bisulfate-based bisulfite conversion method48, 49 and the PCR fragments from the converted DNA was sequenced and analyzed. Raw sequence data files was processed and the area ratio (%) of C over C+T of the primary CpG dinucleotide was calculated as the percenatge of methylation and plotted.50
The in vivo studies
Animal experiments were undertaken in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. BALB/c male nude mice aged of 8–12 weeks were used for this study. 5637 or H-bc cells were embedded in BD Matrigel Matrix (Becton, Dickinson, Franklin Lakes, NJ, USA)51 and subcutaneously injected into the four sites at the back of mice as following: 1.7 × 107 cells/site for 5637, 0.7 × 107 cells/site for H-bc, 4 sites/mouse, 6 mice for 5637, 6 mice for H-bc, respectively. From the fourth day after cell injection, all 5637-generated tumors were intratumorally injected with 2nM miR-193a-3p agomiR or miR-193a-5p agomiR, while H-bc-generated tumors were injected with 4 nM miR-193a-3p/-5p/Mock antagomiR/PBS in a similar manner. From the sixth day after cell injection, three mice from 5637 and three from H-bc intraperitoneally received Pa (45 ug/ mouse) once in 2 days. The remaining six mice (three from 5637 and three from H-bc) received PBS as a mock treatment control. Mice were humanely killed on day 25, and the tumors were weighed and photographed. The tumor weight was described as the mean±S.D.
Expression levels of SRSF2, TP73 and Ki67 proteins were measured using immunochemical analysis on 5-mm slices of formalin-fixed paraffin-embedded tumor xenografts in nude mice. To avoid inter-treatment bias, the tissue slides from all the six groups were made on a single slide and subjected to the same immuno-staining simultaneously. Antigens were retrieved by pretreating dewaxed sections in a microwave oven at 750 Watts for 5 min in a citrate buffer (pH 6) processed with the Super Sensitive Link-Labeled Detection System (Biogenex, Menarini, Florence, Italy). The enzymatic activities were developed using 3-amino-9-ethylcarbazole (Dako, Milan, Italy) as a chromogenic substrate. Following counter staining with Mayer hematoxylin (Invitrogen), slides were mounted in an aqueous mounting medium (glycergel, Dako).Pictures were taken using the LEICA DM 4000B microscope (Wetzlar, Germany), while the relative level of each protein was calculated using the LEICA software (Wetzlar, Germany), and the percentage of the mock over the chemotherapeutic treated tumors was calculated and plotted.
Bioinformatics analysis
R package pheatmap version 0.77 (http://cran.r-project.org/web/packages/pheatmap/index.html) were used to create the diagrams. CpG island information for the human genome (hg19) was obtained from UCSC database.52Differentially expressed miRs were calculated with in-house developed perl scripts. The networking analysis: the following eight genes: PLAU, HIC2, and SRSF2, together with the master transcription factor genes in each pathways (Oxidative stress: RBPJ, MYC/Max: MYC, Notch: NRF1, DNA damage: TP53 and NF−κB pathways: NFKB1) were used as seeds to extract from the known PPI from the STRING database version 9.10.17 The build-in one-step expand algorithm was used until a fully connected network of all seed genes were obtained. To simplify the model, the shortest paths between seeds were extracted from the networks. RNA-seq data (not shown) were used to identify the expression pattern of the model. To obtain the true functional role of each interaction in the connected network of eight seeds and four hub genes, the literature mining was performed by the STRING via full text reading, with both standard gene name and alternative name considered. Edges that are not consistent with our experimental data were removed from the network.
Statistical analysis
Data are presented as means, and error bars indicate the S.D. or S.E. All statistical analyses were performed with Excel (Microsoft, Redmond, WA, USA) or Prism (GraphPad Software Inc., La Jolla, CA, USA). Two-tailed Student’s t-test, a one-way analysis of variance or Mann–Whitney U test was used to calculate statistical significance. A P-value of <0.05 was considered to be significant.
Abbreviations
- BCa:
-
bladder cancer
- MiR:
-
microRNA
- HCC:
-
hepatocellular carcinoma
- Pi:
-
Pirarubicin
- Pa:
-
Paclitaxel
- Ad:
-
Adriamycin
- EH:
-
Epirubicin Hydrochloride
References
Ferlay J, Shin HR, Bray F, Forman D, Mathers C, Parkin DM . Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer 2010; 127: 2893–2917.
Burger M, Catto JW, Dalbagni G, Grossman HB, Herr H, Karakiewicz P et al. Epidemiology and risk factors of urothelial bladder cancer. Eur Urol 2012; 63: 234–241.
von der Maase H, Sengelov L, Roberts JT, Ricci S, Dogliotti L, Oliver T et al. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol 2005; 23: 4602–4608.
Chang JS, Lara PN, Pan C-X . Progress in personalizing chemotherapy for bladder cancer. Adv Urol 2012; 2012: 364919.
Gordon RR, Nelson PS . Cellular senescence and cancer chemotherapy resistance. Drug Resist Updat 2012; 15: 123–131.
Allen KE, Weiss GJ . Resistance may not be futile: microRNA biomarkers for chemoresistance and potential therapeutics. Mol Cancer Ther 2010; 9: 3126–3136.
Vinall RL, Ripoll AZ, Wang S, Pan CX, deVere White RW . MiR-34a chemosensitizes bladder cancer cells to cisplatin treatment regardless of p53-Rb pathway status. Int J Cancer 2012; 130: 2526–2538.
Tao J, Lu Q, Wu D, Li P, Xu B, Qing W et al. microRNA-21 modulates cell proliferation and sensitivity to doxorubicin in bladder cancer cells. Oncol Rep 2011; 25: 1721–1729.
Su S, Chang Y, Andreu-Vieyra C, Fang J, Yang Z, Han B et al. miR-30d, miR-181a and miR-199a-5p cooperatively suppress the endoplasmic reticulum chaperone and signaling regulator GRP78 in cancer. Oncogene 2013; 32: 4694–4701.
Lujambio A, Ropero S, Ballestar E, Fraga MF, Cerrato C, Setién F et al. Genetic unmasking of an epigenetically silenced microRNA in human cancer cells. Cancer Res 2007; 67: 1424–1429.
Brinkman AB, Simmer F, Ma K, Kaan A, Zhu J, Stunnenberg HG et al. Whole-genome DNA methylation profiling using MethylCap-seq. Methods 2010; 52: 232–236.
Ma K, He Y, Zhang H, Fei Q, Niu D, Wang D et al. DNA methylation-regulated miR-193a-3p dictates resistance of hepatocellular carcinoma to 5-fluorouracil via repression of SRSF2 expression. J Biol Chem 2012; 287: 5639–5649.
Iliopoulos D, Rotem A, Struhl K . Inhibition of miR-193a expression by Max and RXRα activates K-Ras and PLAU to mediate distinct aspects of cellular transformation. Cancer Res 2011; 71: 5144–5153.
Noh H, Hong S, Dong Z, Pan ZK, Jing Q, Huang S . Impaired microRNA processing facilitates breast cancer cell invasion by upregulating urokinase-type plasminogen activator expression. Genes Cancer 2011; 2: 140–150.
Salvi A, Conde I, Abeni E, Arici B, Grossi I, Specchia C et al. Effects of miR-193a and sorafenib on hepatocellular carcinoma cells. Mol Cancer 2013; 12: 162.
Ory B, Ramsey MR, Wilson C, Vadysirisack DD, Forster N, Rocco JW et al. A microRNA-dependent program controls p53-independent survival and chemosensitivity in human and murine squamous cell carcinoma. J Clin Invest 2011; 121: 809.
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A et al. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res 2013; 41: D808–D815.
Napolitano G, Mazzocco A, Fraldi A, Majello B, Lania L . Functional inactivation of Cdk9 through oligomerization chain reaction. Oncogene 2003; 22: 4882–4888.
Lin S, Coutinho-Mansfield G, Wang D, Pandit S, Fu X-D . The splicing factor SC35 has an active role in transcriptional elongation. Nat Struct Mol Biol 2008; 15: 819–826.
Baumli S, Lolli G, Lowe ED, Troiani S, Rusconi L, Bullock AN et al. The structure of P-TEFb (CDK9/cyclin T1), its complex with flavopiridol and regulation by phosphorylation. EMBO J 2008; 27: 1907–1918.
Young TM, Wang Q, Pe'ery T, Mathews MB . The human I-mfa domain-containing protein, HIC, interacts with cyclin T1 and modulates P-TEFb-dependent transcription. Mol Cell Biol 2003; 23: 6373–6384.
Yong FL, Law CW, Wang CW . Potentiality of a triple microRNA classifier: miR-193a-3p, miR-23a and miR-338-5p for early detection of colorectal cancer. BMC Cancer 2013; 13: 280.
Heller G, Weinzierl M, Noll C, Babinsky V, Ziegler B, Altenberger C et al. Genome-wide miRNA expression profiling identifies miR-9-3 and miR-193a as targets for DNA methylation in non–small cell lung cancers. Clin Cancer Res 2012; 18: 1619–1629.
Gao X, Lin J, Li Y, Gao L, Wang X, Wang W et al. MicroRNA-193a represses c-kit expression and functions as a methylation-silenced tumor suppressor in acute myeloid leukemia. Oncogene 2011; 30: 3416–3428.
Watson JA, Bryan K, Williams R, Popov S, Vujanic G, Coulomb A et al. miRNA profiles as a predictor of chemoresponsiveness in Wilms’ tumor blastema. PLoS One 2013; 8: e53417.
Takeshita H, Shiozaki A, Bai X-H, Iitaka D, Kim H, Yang BB et al. XB130, a new adaptor protein, regulates expression of tumor suppressive MicroRNAs in cancer cells. PLoS One 2013; 8: e59057.
Wang J, Yang B, Han L, Li X, Tao H, Zhang S et al. Demethylation of miR-9-3 and miR-193a genes suppresses proliferation and promotes apoptosis in non-small cell lung cancer cell lines. Cell Physiol Biochem 2013; 32: 1707–1719.
Nakano H, Yamada Y, Miyazawa T, Yoshida T . Gain-of-function microRNA screens identify miR-193a regulating proliferation and apoptosis in epithelial ovarian cancer cells. Int J Oncol 2013; 42: 1875.
Chekhun VF, Kulik GI, Yurchenko OV, Tryndyak VP, Todor IN, Luniv LS et al. Role of DNA hypomethylation in the development of the resistance to doxorubicin in human MCF-7 breast adenocarcinoma cells. Cancer Lett 2006; 231: 87–93.
Alfano D, Iaccarino I, Stoppelli MP . Urokinase signaling through its receptor protects against anoikis by increasing BCL-xL expression levels. J Biol Chem 2006; 281: 17758–17767.
Green WB, McGuire PG, Miska KB, Kusewitt DF . Urokinase activity in corneal fibroblasts may be modulated by DNA damage and secreted proteins. Photochem Photobiol 2001; 73: 318–323.
Bittel D, Yu S, Newkirk H, Kibiryeva N, Holt Iii A, Butler M et al. Refining the 22q11. 2 deletion breakpoints in DiGeorge syndrome by aCGH. Cytogenet Genome Res 2009; 124: 113–120.
Han J-W, Zheng H-F, Cui Y, Sun L-D, Ye D-Q, Hu Z et al. Genome-wide association study in a Chinese Han population identifies nine new susceptibility loci for systemic lupus erythematosus. Nat Genet 2009; 41: 1234–1237.
Cazalla D, Zhu J, Manche L, Huber E, Krainer AR, Cáceres JF . Nuclear export and retention signals in the RS domain of SR proteins. Mol Cell Biol 2002; 22: 6871–6882.
Edmond V, Moysan E, Khochbin S, Matthias P, Brambilla C, Brambilla E et al. Acetylation and phosphorylation of SRSF2 control cell fate decision in response to cisplatin. EMBO J 2010; 30: 510–523.
Li Y, Kimura T, Huyck RW, Laity JH, Andrews GK . Zinc-induced formation of a coactivator complex containing the zinc-sensing transcription factor MTF-1, p300/CBP, and Sp1. Mol Cell Biol 2008; 28: 4275–4284.
Kurooka H, Honjo T . Functional interaction between the mouse notch1 intracellular region and histone acetyltransferases PCAF and GCN5. J Biol Chem 2000; 275: 17211–17220.
Izumi H, Ohta R, Nagatani G, Ise T, Nakayama Y, Nomoto M et al. p300/CBP-associated factor (P/CAF) interacts with nuclear respiratory factor-1 to regulate the UDP-N-acetyl-alpha-d-galactosamine: polypeptide N-acetylgalactosaminyltransferase-3 gene. Biochem J 2003; 373: 713–722.
Kanazawa S, Soucek L, Evan G, Okamoto T, Peterlin BM . c-Myc recruits P-TEFb for transcription, cellular proliferation and apoptosis. Oncogene 2003; 22: 5707–5711.
Redwood SM, Liu BC, Weiss RE, Hodge DE, Droller MJ . Abrogation of the invasion of human bladder tumor cells by using protease inhibitor (s). Cancer 1992; 69: 1212–1219.
Xu B, He Y, Wu X, Luo C, Liu A, Zhang J . Exploration of the correlations between interferon-gamma in patient serum and HEPACAM in bladder transitional cell carcinoma, and the interferon-gamma mechanism inhibiting BIU-87 proliferation. J Urol 2012; 188: 1346–1353.
El-Sheikh A, Fan R, Birks D, Donson A, Foreman NK, Vibhakar R . Inhibition of aurora kinase A enhances chemosensitivity of medulloblastoma cell lines. Pediatr Blood Cancer 2010; 55: 35–41.
Xu L, Wang F, Xu X-F, Mo W-H, Xia Y-J, Wan R et al. Down-regulation of miR-212 expression by DNA hypermethylation in human gastric cancer cells. Med Oncol 2011; 28: 189–196.
Li J, Chen F, Cona MM, Feng Y, Himmelreich U, Oyen R et al. A review on various targeted anticancer therapies. Target Oncol 2012; 7: 69–85.
Heiser LM, Sadanandam A, Kuo WL, Benz SC, Goldstein TC, Ng S et al. Subtype and pathway specific responses to anticancer compounds in breast cancer. Proc Natl Acad Sci USA 2012; 109: 2724–2729.
Andrisano V, Bartolini M, Gotti R, Cavrini V, Felix G . Determination of inhibitors' potency (IC50) by a direct high-performance liquid chromatographic method on an immobilised acetylcholinesterase column. J Chromatogr B Biomed Sci Appl 2001; 753: 375–383.
Song B, Wang Y, Xi Y, Kudo K, Bruheim S, Botchkina GI et al. Mechanism of chemoresistance mediated by miR-140 in human osteosarcoma and colon cancer cells. Oncogene 2009; 28: 4065–4074.
Shiraishi M, Hayatsu H . High-speed conversion of cytosine to uracil in bisulfite genomic sequencing analysis of DNA methylation. DNA Res 2004; 11: 409–415.
Hayatsu H, Negishi K, Shiraishi M . DNA methylation analysis: speedup of bisulfite-mediated deamination of cytosine in the genomic sequencing procedure. Proc Jpn Acad B 2004; 80: 189–194.
Lewin J, Schmitt AO, Adorján P, Hildmann T, Piepenbrock C . Quantitative DNA methylation analysis based on four-dye trace data from direct sequencing of PCR amplificates. Bioinformatics 2004; 20: 3005–3012.
Ohashi K, Marion PL, Nakai H, Meuse L, Cullen JM, Bordier BB et al. Sustained survival of human hepatocytes in mice: a model for in vivo infection with human hepatitis B and hepatitis delta viruses. Nat Med 2000; 6: 327–331 PubMed PMID: 10700236. Epub 2000/03/04. eng.
Bock C, Walter J, Paulsen M, Lengauer T . CpG island mapping by epigenome prediction. PLoS Comput Biol 2007; 3: e110.
Acknowledgements
This work was supported by The 973 National Key Fundamental Research Program of China (2010CB912802 to JZ), the National Natural Science Foundation of China (30921140312 to JZ, 81171996 and 81272289 granted to CZ, 81200975 granted to YL), the cooperational projects M011, MHBI (shanghai) Biotech (to XL), the Anhui Provincial Natural Science Foundation (1208085QH166 to YL) and Wujieping Medical Foundation ( No.320.6750.13252 to CZ).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Dr. Xiao Liu is the founder of MHBI (Shanghai) Biotechnology Incorporation. The other authors declare no conflict of interest.
Additional information
Edited by G Calin
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution-NonCommercial-ShareAlike 3.0 Unported License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-sa/3.0/
About this article
Cite this article
Lv, L., Deng, H., Li, Y. et al. The DNA methylation-regulated miR-193a-3p dictates the multi-chemoresistance of bladder cancer via repression of SRSF2/PLAU/HIC2 expression. Cell Death Dis 5, e1402 (2014). https://doi.org/10.1038/cddis.2014.367
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.367
This article is cited by
-
Noncoding RNAs regulate alternative splicing in Cancer
Journal of Experimental & Clinical Cancer Research (2021)
-
HSD17B6 downregulation predicts poor prognosis and drives tumor progression via activating Akt signaling pathway in lung adenocarcinoma
Cell Death Discovery (2021)
-
Non coding RNAs as the critical factors in chemo resistance of bladder tumor cells
Diagnostic Pathology (2020)
-
Low doses of decitabine improve the chemotherapy efficacy against basal-like bladder cancer by targeting cancer stem cells
Oncogene (2019)
-
Paraoxonase 3 is involved in the multi-drug resistance of esophageal cancer
Cancer Cell International (2018)