Abstract
Mesenchymal stem cells (MSCs) are able to infiltrate tumor tissues and thereby effectively deliver gene therapeutic payloads. Here, we engineered murine MSCs (mMSCs) to express a secreted form of the TNF-related apoptosis-inducing ligand (TRAIL), which is a potent inducer of apoptosis in tumor cells, and tested these MSCs, termed MSC.sTRAIL, in combination with conventional chemotherapeutic drug treatment in colon cancer models. When we pretreated human colorectal cancer HCT116 cells with low doses of 5-fluorouracil (5-FU) and added MSC.sTRAIL, we found significantly increased apoptosis as compared with single-agent treatment. Moreover, HCT116 xenografts, which were cotreated with 5-FU and systemically delivered MSC.sTRAIL, went into remission. Noteworthy, this effect was protein 53 (p53) independent and was mediated by TRAIL-receptor 2 (TRAIL-R2) upregulation, demonstrating the applicability of this approach in p53-defective tumors. Consequently, when we generated MSCs that secreted TRAIL-R2-specific variants of soluble TRAIL (sTRAIL), we found that such engineered MSCs, labeled MSC.sTRAILDR5, had enhanced antitumor activity in combination with 5-FU when compared with MSC.sTRAIL. In contrast, TRAIL-resistant pancreatic carcinoma PancTu1 cells responded better to MSC.sTRAILDR4 when the antiapoptotic protein XIAP (X-linked inhibitor of apoptosis protein) was silenced concomitantly. Taken together, our results demonstrate that TRAIL-receptor selective variants can potentially enhance the therapeutic efficacy of MSC-delivered TRAIL as part of individualized and tumor-specific combination treatments.
Similar content being viewed by others
Main
The tumor necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) is a potent and specific inducer of apoptosis in tumor cells and can also act as a powerful cancer-preventive agent.1, 2 It can bind to four membrane-bound receptors and one soluble receptor (TRAIL-R1, TRAIL-R2, TRAIL-R3, TRAIL-R4 and osteoprotegerin also known as DR4, DR5, decoy receptor 1 (DcR1), decoy receptor 2 (DcR2) and OPG, respectively). Both TRAIL receptors 1 and 2 (TRAIL-R1 and TRAIL-R2) contain a conserved cytoplasmic region termed the death domain (DD) that is required for TRAIL-induced apoptosis.3 Upon binding of TRAIL, a protein complex is formed known as death-inducing signaling complex (DISC) consisting of TRAIL-R1 and/or TRAIL-R2, the adaptor protein Fas-associated death domain (FADD) and procaspase-8.4 Activation of caspase-8 at the DISC and subsequent activation of effector caspases leads to apoptosis.5 In contrast, TRAIL receptors 3 and 4 (TRAIL-R3 and TRAIL-R4) and the soluble receptor OPG have inhibitory functions, because they lack the intracellular DDs, and are therefore regarded as decoy receptors.1 Therefore, one potential way of improving the therapeutic efficacy of TRAIL is to generate agents that act in a TRAIL-R-specific way, preferring one of the two death receptors and potentially avoiding the decoy receptors. To this end, agonistic antibodies directed against one of the two apoptosis-inducing TRAIL-Rs have been developed and tested in various experimental systems as well as in the clinic.6, 7, 8, 9 In addition, engineered variants of the TRAIL protein, containing specific amino-acid changes leading to specific targeting of TRAIL-R1 or TRAIL-R2 have been developed and have shown enhanced antitumor properties in vitro and in vivo.10, 11, 12, 13 However, in clinical trials with recombinant TRAIL (rTRAIL) protein and agonistic antibodies to TRAIL-R2, only moderate therapeutic activities have been found.14 A possible explanation for the problems with recombinant protein and antibodies is limited bioavailability owing to short half-life of the protein or restricted accessibility of large antibodies to tumor tissue, respectively.15 Cell and gene therapy approaches provide a potential strategy to overcome such limitations and several cellular gene delivery vectors utilizing TRAIL as its therapeutic payload have shown efficacy in experimental tumor models.16, 17, 18, 19, 20 In particular, mesenchymal stem cells (MSCs) have been used in this context, as they are known to readily infiltrate tumor tissues.21, 22 However, limitations to this approach have been highlighted in a recent study that demonstrated the requirement for substantial intratumoral presence of MSCs to afford growth inhibition in a colorectal carcinoma model.23 Therefore, it is essential to derive as much therapeutic effect from the tumor-infiltrating MSCs as possible, because in a clinical setting the number of MSCs that can be safely injected and consequently the number of tumor-infiltrating MSCs might be limited. Thus, the use of sTRAIL as compared with full-length membrane-bound TRAIL19 and integration of TRAIL-R-specific variants10, 11, 12, 13 are valuable approaches to augment the antineoplastic effects of the MSC-based delivery system. A further barrier for TRAIL therapies is that a substantial proportion of tumors are relatively resistant to the effects of TRAIL.24 To this end, we could show that a sensitization approach with the commonly used anticancer compound 5-fluorouracil (5-FU) in conjunction with systemically administered MSC.sTRAIL worked in vitro and in vivo. The effect was mediated by protein 53 (p53)-independent increases in the expression of TRAIL-R2, and consequently MSCs secreting a TRAIL-R2-specific sTRAIL variant (MSC.sTRAILDR5) gave rise to significantly higher apoptosis than MSC.sTRAIL (wild-type sTRAIL) or MSC.sTRAILDR4 (TRAIL-R1-specific variant) in 5-FU-pretreated HCT116 cells. In a second sensitization approach, we targeted the antiapoptotic protein XIAP (X-linked inhibitor of apoptosis protein) by RNA interference (RNAi) in TRAIL-resistant pancreatic carcinoma cells and could show that MSC.sTRAILDR4 exerted significantly increased apoptosis-inducing activity as compared with MSC.sTRAIL or MSC.sTRAILDR5. Hence, MSC-mediated delivery of optimized sTRAIL variants to tumors combined with conventional chemotherapy or inhibition of apoptosis resistance factors such as XIAP could become a novel, patient-tailored treatment strategy for various oncological diseases.
Results
Combined treatment of 5-FU and TRAIL leads to synergistic apoptosis induction
Because TRAIL as a stand-alone strategy is unlikely to be efficacious in all cancer types and cases owing to apoptosis resistance of tumor cells, we set out to test optimized TRAIL-based approaches utilizing combination treatments with the cytotoxic drug 5-FU and MSC-mediated delivery of sTRAIL. First, we determined the minimal concentrations of rTRAIL and 5-FU that could be used in a combination treatment of colorectal cancer cells. HCT116 cells were treated with 5-FU at concentrations of 10, 50, 100, 200 and 400 μM (Figure 1a) and rTRAIL at concentrations of 0.25, 0.5, 1, 2 and 5 ng/ml (Figure 1a). The results show increasing apoptosis levels with rising concentrations of the respective apoptosis-inducing agent. For our combination treatment experiment, we chose concentrations of the two agents that gave rise to relatively low specific apoptosis levels and thus pretreated HCT116 cells with 10 μM 5-FU for 24 h followed by 1 ng/ml rTRAIL for an additional 24 h before we measured cell death. This combined treatment resulted in 37±0.6% apoptosis as compared with 2.6±0.4% and 10.5±0.4% apoptosis, respectively, for the single treatments (Figure 1a). Apoptosis could be completely blocked by the pan-caspase inhibitor zVAD (carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone), demonstrating that the detected cell death was a caspase-dependent apoptotic process (Figure 1a).
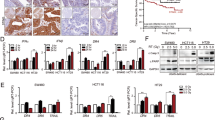
Cancer cells can be sensitized to the effects of MSC.sTRAIL by 5-FU. (a) Apoptosis was measured after exposure of HCT116 cells to increasing doses of 5-FU. Cells were treated for 48 h with the respective doses of 5-FU (left). HCT116 cells were treated with rTRAIL with increasing concentrations and apoptosis measured 24 h later (center). Apoptosis measurements after treatment with 10 μM 5-FU for 48 h, treatment with rTRAIL at 1 ng/ml for 24 h or pretreatment with 10 μM 5-FU for 24 h followed by 1 ng/ml rTRAIL for an additional 24 h. Addition of zVAD directly to the HCT116 cells before 5-FU pretreatment completely inhibited apoptosis of the combined treatment (5-FU followed by rTRAIL). Carrier (dimethylsulfoxide (DMSO)) treatment had no effect (right). Data represent averages±S.E.M. (n=3, P<0.001). (b) Schematic drawing of the sTRAIL construct depicts the signal peptide sequence from the human Fibrillin-1 gene (hFIB) that was ligated to the Furin cleavage site (Furin CS), an Isoleucine zipper (ILZ) stretch and the sTRAIL (ectodomain) part (aa114–aa289). The expression was driven by a conventional CMV promoter/enhancer element (left). Murine MSCs and their differentiation potential was analyzed by a von-Kossa osteogenesis assay, an Oil Red O staining for adipogenic differentiation and Toluidine Blue staining for chondrogenesis (center). FACS analyses of mouse MSCs for surface expression of CD117, CD11b, CD19, CD45, TER-119, CD44, Sca-1 and CD29. The three respective isotype controls are shown in gray (right). (c) Mixing of 10 μM 5-FU-pretreated HCT116 cells with MSC.sTRAIL in a ratio of 10 : 1 showed significantly increased apoptosis as compared with 5-FU/MSC.DsRed or MSC.sTRAIL treatments. Additional controls are 5-FU and MSC.DsRed treatments (left). Data represent averages±S.E.M. (n=3, P<0.001). Transfer of medium supernatants (SNs) from MSC.sTRAIL onto 5-FU-pretreated HCT116 cells also gave rise to significantly higher apoptosis levels compared with SNs from MSC.sTRAIL that were applied to non-5-FU-treated HCT116 cells or in 5-FU/MSC.DsRed samples. Additional controls are supernatants from MSC.DsRed or 5-FU-only treated HCT116 cells (right). Data represent averages±S.E.M. (n=3, P<0.001)
A significant problem for clinical use of TRAIL is effective delivery and sufficient bioavailability in tumor lesions of the recombinant protein or agonistic TRAIL-R antibodies.15 In order to overcome this problem, we and others have exploited the tumor-infiltrating properties of MSCs to deliver sTRAIL.17, 18, 19 Here, we have constructed an adenoviral (Ad) vector (Ad.sTRAIL) that contained the ectodomain of TRAIL (aa114–aa289) fused to the secretory signal peptide from the human Fibrillin-1 gene (hFIB), a Furin cleavage site (Furin CS) and an Isoleucine zipper (ILZ) domain under the control of the cytomegalovirus (CMV) promoter/enhancer element (Figure 1b).25 The vector Ad.sTRAIL was used to transduce MSCs, which had been characterized by their capacity for adipogenic, osteogenic and chondrogenic differentiation (Figure 1b) as well as by fluorescence-activated cell sorting (FACS) analyses with an array of surface marker antibodies. The results show that our murine MSCs (mMSCs) are CD29+, Sca-1+, CD44+, CD11b−, CD19−, CD117−, CD45− and Ter−119−, demonstrating their purity and ruling out a contamination with fibroblasts and hematopoietic cells (Figure 1b). The resulting modified MSCs that expressed and secreted sTRAIL were termed MSC.sTRAIL (Supplementary Figure S1a–d). Mixing of MSC.sTRAIL with HCT116 cells that had been pretreated with 10 μM 5-FU for 24 h gave rise to significant increased apoptosis levels in the double-treated cells (5-FU/MSC.sTRAIL) as compared with the single treatments or a 5-FU/MSC.DsRed control combination (Figure 1c). As we obtained the same significant sensitization effect when we transferred the medium supernatant from MSC.sTRAIL 48 h after transduction with Ad.sTRAIL onto 5-FU-pretreated HCT116, we could conclude that the effect was indeed exerted by secreted sTRAIL and not via direct MSC–cancer cell interactions (Figure 1c).
Combined treatment of 5-FU and TRAIL is efficacious and safe in vivo
Based on our in vitro findings we examined the utility of 5-FU in combination with MSC.sTRAIL in HCT116 xenografts. First, we tested the duration of transgene expression in adenovirally transduced MSCs in vitro to inform our in vivo regimen (Supplementary Figure S2) and assessed the effect of 5-FU on MSCs and their potential to secrete sTRAIL as well as to induce apoptosis in the presence of 5-FU. The results revealed that a second injection of MSC.sTRAIL 10 days after the first administration might be helpful, as transgene expression in MSCs dropped substantially between day 8 and day 12 after transduction and that MSCs are 5-FU resistant and sTRAIL secretion is not affected by 5-FU (Supplementary Figure S3a–d). Thus, MSCs can be used as cellular delivery vehicle in the context of an experimental 5-FU/MSC.sTRAIL treatment.
Next, we established tumors in immune-deficient nu/nu mice. These mice were then intraperitoneally injected with 150 mg/kg 5-FU before 1 × 105 MSC.sTRAIL were systemically administered via the tail vein. After 10 days, the mice were treated with a second dose of 1 × 105 MSC.sTRAIL. As controls, we tested tumors that were treated with MSC.DsRed together with 5-FU as well as MSC.sTRAIL or MSC.DsRed alone. Although the tumors in the control group, treated with MSC.DsRed, grew almost exponentially, xenografts either treated with 5-FU/MSC.DsRed or MSC.sTRAIL showed marked growth reduction. Most strikingly, tumors treated with the combination of 5-FU and MSC.sTRAIL went into remission (Figure 2a). When we analyzed the tumors histologically by hematoxylin and eosin (H&E) staining to examine general tissue morphology, and by Masson’s trichrome staining to visualize the connective tissue (collagen fibers), we found MSC.DsRed and 5-FU/MSC.DsRed sections showing a nonencapsulated tumor with cancer cell infiltration of the surrounding muscle tissue in the H&E analysis (Figure 2b). The same tumors stained with the Masson’s trichrome method showed that 5-FU/MSC.DsRed had some fiber development inside the tumor mass (Figure 2b). H&E- and Masson’s trichrome-stained MSC.sTRAIL tumor samples showed fiber formation surrounding the tumor that still looked proliferating but limited by a capsule (Figure 2b). In contrast, 5-FU/MSC.sTRAIL clearly showed a lot of cellular debris mixed with collagen fibers replacing the proliferating cells that were present in the other samples (Figure 2b). In addition, nuclear proliferating cell nuclear antigen (PCNA) protein expression, which is observed during DNA synthesis and generally represents cellular proliferating activity, was detected immunohistochemically (Figure 2b). In the MSC.DsRed and 5-FU/MSC.DsRed groups, PCNA levels were higher compared with MSC.sTRAIL tumor samples and were almost absent in sections from the 5-FU/MSC.sTRAIL group (Figure 2b). Hence, whereas 5-FU and MSC.sTRAIL as single-agent regimens possess significant but limited anticancer activities, the combination of both gave rise to tumor remission.
Treatment with 5-FU and MSC.sTRAIL lead to tumor remission in vivo. (a) Immune-deficient mice were injected with 5 × 106 HCT116 cells. After 10 days (= day 0 of treatment regimen) 150 mg/kg 5-FU was injected intraperitoneally and subsequently 1 × 105 MSC.DsRed (blue line; n=4) or MSC.sTRAIL (red line; n=6) was intravenously administered. Animals in two additional control groups received MSC.DsRed (black line; n=4) or MSC.sTRAIL (green line; n=4) alone without 5-FU. The animals were injected with engineered MSCs on day 0 and again on day 10, as indicated by arrows. The tumor growth was followed over 33 days and is depicted in the graph (P=0.003, P=0.02 and P=0.03). (b) H&E staining (upper panel) to examine general tumor tissue morphology and Masson’s trichrome staining (center panel) to visualize the connective tissue (collagen in connective tissue is stained blue and cytoplasm appears in pink) on microsections from MSC.DsRed-, 5-FU/MSC.DsRed-, MSC.sTRAIL- and 5-FU/MSC.sTRAIL-treated HCT116 tumor-burdened animals. Immunohistochemical detection of nuclear PCNA protein expression (brown color) on microsections of MSC.DsRed-, 5-FU/MSC.DsRed-, MSC.sTRAIL- and 5-FU/MSC.sTRAIL-treated HCT116 tumors (lower panel). Scale bars, 200 μm. (c) Apoptosis was measured after exposure of murine and human MSCs to 10 μM 5-FU for 48 h (black bars), treatment with rTRAIL at 10 ng/ml for 24 h (red bars) or pretreatment with 10 μM 5-FU for 24 h followed by 10 ng/ml rTRAIL for an additional 24 h (white bars). HCT116 cells treated with the same concentration of 5-FU, but with 1 ng/ml rTRAIL, served as positive control. Data represent averages±S.E.M. (n=3, P<0.001). (d) H&E staining of liver sections from mice treated with MSC.DsRed, 5-FU/MSC.DsRed, MSC.sTRAIL and 5-FU/MSC.sTRAIL, respectively. Scale bars, 500 μm. High-power images of the boxed-in areas of the respective tissue sections are shown in the lower panel of images. Scale bars, 200 μm
Next, we tested the safety of combined 5-FU/TRAIL treatments on primary murine and human cells. To this end, we incubated murine and human MSCs with 10 μM 5-FU for 24 h followed by rTRAIL (10 ng/ml) for an additional 24 h period. When we measured apoptosis in these cells, we found that they were resistant to the single treatments as well as to the combination of 5-FU and rTRAIL (Figure 2c). Furthermore, we examined the livers of mice that had received 5-FU injections followed by MSC.sTRAIL and could not detect any signs of tissue injury or cell death in these tissue samples (Figure 2d), indicating that the present novel therapeutic approach is safe.
The molecular mechanism of TRAIL sensitization by 5-FU
In order to further improve TRAIL-based combination treatments, a better understanding of the underlying molecular mechanism(s) of the observed sensitization is important. First, we analyzed caspase-8 activation, which is one of the initial molecular events in the TRAIL-triggered apoptosis pathway, in 5-FU/TRAIL-treated HCT116 cells. Our results demonstrate that caspase-8 activation is markedly increased in cells exposed to the combined treatment as compared with cells treated with either 5-FU or rTRAIL alone (Figure 3a). In order to show that the 5-FU sensitization effect is indeed transduced through the canonical caspase-8-initiated apoptosis pathway or whether additional, independent mechanisms and factors are required, we generated stable caspase-8 knockdowns in HCT116 cells, termed HCT.shc8 (Figure 3a). These cells were treated with the standard 5-FU/TRAIL regimen and apoptosis was measured showing that in HCT.shc8 cells apoptosis was completely inhibited, demonstrating that caspase-8 was the sole initiator caspase and that the sensitization effect of 5-FU was not mediated by mechanisms outside of the normal extrinsic apoptosis pathway (Figure 3a). Thus, molecularly, the observed 5-FU-mediated sensitization is located at the level of caspase-8 activation or upstream of it. Although these results show that caspase-8 is required for 5-FU/TRAIL-triggered apoptosis, we could not detect any indications that 5-FU gave rise to elevated levels of caspase-8, thereby making a role as molecular driver of the observed 5-FU sensitization effect unlikely. Instead, we turned to the antiapoptotic protein cFlip (cellular FLICE-like inhibitory protein), as others have implicated 5-FU-triggered downregulation of cFlip in the sensitization effect.26 However, although decreased levels of cFlip protein after 5-FU stimulation could indeed explain enhanced caspase-8 activation, we were unable to detect any decreases in cFlip levels (Figure 3b). Next, we examined changes in the surface levels of TRAIL-R1 and TRAIL-R2 by FACS after 5-FU treatment, because increased TRAIL-R expression could lead to higher caspase-8 activation and apoptosis in response to TRAIL, and as in particular TRAIL-R2 has been shown to be upregulated by 5-FU via a mechanism that is believed to be p53 dependent.27 Following 5-FU treatment, we detected a substantial rise in TRAIL-R2 expression but almost no effect on the TRAIL-R1 levels (Figure 3b). To test whether the increased TRAIL-R2 levels could prime the cells for increased TRAIL responsiveness, we generated stable TRAIL-R2 and as controls TRAIL-R1 knockdown clones in HCT116 cells, labeled HCT.shDR5 and HCT.shDR4, respectively (Figure 3c). When the HCT.shDR4 cells were treated with 5-FU followed by rTRAIL, we observed significantly elevated levels of cell death as compared with control cells (Figure 3c). In contrast, in HCT.shDR5 cells, 5-FU/TRAIL-induced apoptosis was significantly reduced (Figure 3c). These results strongly point to the upregulation of TRAIL-R2 as the molecular driver of the 5-FU-induced sensitization effect. In contrast, TRAIL-R1 not only had no role in the observed sensitization, but silencing of this receptor gave rise to a significant increase in 5-FU/TRAIL-induced apoptosis, most likely owing to the fact that TRAIL-R2 molecules can form more readily active homotrimers and consequently induce a higher level of apoptosis with no or very little interfering TRAIL-R1 present in the membrane. Following these findings, we wondered whether the upregulation of TRAIL-R2 and consequent sensitization to TRAIL is p53 dependent. When we subjected HCT116 and HCTp53−/− cells to the standard 5-FU pretreatment and measured TRAIL-R2 and TRAIL-R1 levels, we found TRAIL-R2 protein expression to be upregulated in HCTp53−/− cells 24 h later as compared with HCT116 cells (Figure 3d), whereas TRAIL-R1 levels were almost unchanged in both cell types. Consequently, 5-FU-mediated TRAIL-induced apoptosis sensitization reached the same levels in HCTp53−/− cells as in HCT116 cells, although in a time-delayed fashion (Figure 3d). To test that 5-FU sensitization is not restricted to HCT116 and HCTp53−/− cells, we additionally analyzed HT-29 human colorectal cancer cells, which harbor a R273H p53 mutation, and found that they could also be sensitized to the apoptosis-inducing effects of TRAIL by pretreatment with 5-FU (Figure 3d). These results demonstrate that the 5-FU-mediated sensitization strategy is independent of p53, but based on p53 status, modified regimens have to be used with respect to the optimal time point of the TRAIL or MSC.sTRAIL administration. Furthermore, given the importance of TRAIL-R2, we hypothesized that the use of TRAIL-R-specific variants that specifically induce apoptosis via TRAIL-R2 might increase the efficacy of the MSC.sTRAIL system further.
5-FU-induced, p53-independent TRAIL-R2 upregulation is responsible for the sensitization effect. (a) Caspase-8 western blot of HCT116 cells that were either left untreated, treated with 5-FU (10 μM; 48 h), rTRAIL (1 ng/ml; 24 h) or 5-FU (10 μM; 24 h) followed by rTRAIL (1 ng/ml; additional 24 h). Pro-caspase bands are marked by an (*), whereas cleavage products indicative of activation are labeled by arrows. The CuZn superoxide dismutase (CuZnSOD) blot is a loading control (left). Caspase-8 western blot of protein lysates from a control knockdown clone of HCT116 cells (HCT.shctrl) and a caspase-8 knockdown clone (HCT.shc8). The CuZnSOD blot served as loading control (center). HCT.shctrl (black bars) and HCT.shc8 (red bars) were pretreated with 5-FU (10 μM) for 24 h and then treated with rTRAIL (1 ng/ml) for 24 h before apoptosis was measured. Controls include HCT.shctrl and HCT.shc8 cells treated only with 5-FU or rTRAIL (right). Data represent averages±S.E.M. (n=3, P<0.001). (b) A cFlip western blot of protein lysates from untreated and 5-FU-stimulated (10 μM; 48 h) HCT116 cells. The CuZnSOD immunoblot is the loading control (left). FACS analyses for TRAIL-R1 and TRAIL-R2 demonstrated that only TRAIL-R2 is substantially upregulated following treatment with 5-FU for 24 h (black bars) and 48 h (green bars) (right). (c) Western blots for TRAIL-R1 (top panel) and TRAIL-R2 (lower panel) of protein lysates from HCT.shctrl clones as well as from HCT116 derivatives with RNAi-mediated silenced TRAIL-R1 (HCT.shDR4) and TRAIL-R2 (HCT.shDR5), respectively. CuZnSOD western blots served as loading controls (left). HCT.shDR4 (red bars) and HCT.shctrl cells (black bars) were pretreated with 5-FU (10 μM) for 24 h followed by rTRAIL (1 ng/ml) for an additional 24 h and apoptosis was measured. Controls include HCT.shctrl and HCT.shDR4 cells treated with 5-FU or rTRAIL alone (center). Data represent averages±S.E.M. (n=3, P<0.001). HCT.shDR5 (red bars) and HCT.shctrl (black bars) were pretreated with 5-FU for 24 h and then stimulated with rTRAIL (1 ng/ml) for an additional 24 h and apoptosis was measured. Additional controls were HCT.shctrl and HCT.shDR5 cells treated with 5-FU or rTRAIL alone (right). Data represent averages±S.E.M. (n=3, P<0.001). (d) FACS analyses of fold upregulation of surface expression of TRAIL-R1 (left) and TRAIL-R2 (center left) in HCT116 and HCTp53−/− cells at 24 h (black bars), 48 h (green bars) and 72 h (blue bars) after 5-FU treatment (10 μM) in relation to untreated cells. Apoptosis measurements in HCTp53−/− cells after pretreatment with 10 μM 5-FU for 24 h, followed by treatment with rTRAIL at 1 ng/ml for an additional 24 h (black bars) and pretreatment with 10 μM 5-FU for 48 h followed by 1 ng/ml rTRAIL for an additional 24 h (green bars) (center right). Data represent averages±S.E.M. (n=3, P<0.001). HT-29 cells were treated with 10 μM 5-FU for 72 h, treated with rTRAIL at 0.5 ng/ml for 24 h and pretreated with 10 μM 5-FU for 48 h followed by 0.5 ng/ml rTRAIL for an additional 24 h, respectively (right). Data represent averages±S.E.M. (n=3, P<0.001)
Soluble TRAIL variants sTRAILDR5 and sTRAILDR4 are specific for TRAIL-R2 and TRAIL-R1, respectively
In addition to the wild-type TRAIL construct containing the consensus TRAIL cDNA sequence, we integrated two TRAIL-R-specific variants, one selective for TRAIL-R1 (DR4) and one for TRAIL-R2 (DR5) into our expression construct. These variants were designed by using the automatic design algorithm FOLD-X28 and have been previously described and tested as recombinant proteins (without the ILZ).13 One amino-acid sequence change was introduced into the TRAIL segment, namely an aspartate to histidine change at position 269 (D269H) to generate the DR5-specific variant (sTRAILDR5), whereas the DR4-specific variant (sTRAILDR4) was generated by a serine to argenine change at position 159 (S159R) (Figure 4a). The resulting expression constructs were transfected into 293 cells, tested by Western blot (Figure 4b) and the level of sTRAIL in the supernatant measured by ELISA (Figure 4b). The results demonstrate that the different sTRAIL variants were expressed and secreted at comparable levels. In order to confirm the TRAIL receptor specificity of the sTRAILDR5 and sTRAILDR4 variants, we used the HCT.shDR5 and HCT.shDR4 cells. When these cells were treated with supernatants from 293 cells secreting sTRAILDR5 or sTRAILDR4, we found that HCT.shDR5 cells showed markedly reduced apoptosis in response to sTRAILDR5 in comparison with HCT116 cells, whereas the cell death levels with sTRAILDR4 and sTRAIL were normal (Figure 4c). In contrast, HCT.shDR4 exhibited significantly decreased apoptosis with sTRAILDR4, but elevated levels with sTRAILDR5 (Figure 4c). These findings indicate that the sTRAIL variants indeed induce apoptosis via one of the two receptors in a specific manner and that they hold promise to improve the effectiveness of our MSC.sTRAIL system.
Receptor-specific sTRAIL variants are expressed and act specifically. (a) Schematic drawing of the sTRAIL, sTRAILDR5 (TRAIL-R2-specific) and sTRAILDR4 (TRAIL-R1-specific) constructs. The TRAILR-specific variants were generated by site-directed mutagenesis inserting a D269H and an S159R amino-acid exchange in the sTRAIL ectodomain, respectively. The expression was driven by a conventional CMV promoter/enhancer element in all three constructs. (b) These sTRAIL expression constructs were transiently transfected into 293 cells. After 24 h, whole-cell lysates were analyzed by western blotting for TRAIL, revealing similar expression levels for all three constructs. CuZn superoxide dismutase (CuZnSOD) was used as a loading control (left). Results of ELISA analyses for TRAIL demonstrating similar secretion of sTRAIL (black), sTRAILDR5 (green) and sTRAILDR4 (red) into the supernatant of transfected 293 cells. Results for cells transfected with an enhanced green fluorescent protein (EGFP) control plasmid are shown in gray (right). (c) Supernatants from 293 cells transfected with either an EGFP control expression plasmid (gray), sTRAIL (black), sTRAILDR5 (green) or sTRAILDR4 (red) were normalized to 2 ng/ml TRAIL (the EGFP control was diluted 1 : 2 in fresh medium) and then applied to HCT116, HCT.shDR5 and HCT.shDR4 cells, respectively, before apoptosis was measured 24 h later. Data represent averages±S.E.M. (n=3, P=0.008, P=0.01, P=0.004)
The variants sTRAILDR5 and sTRAILDR4 induce increased apoptosis in different cancer cells
To further test the activities of the TRAIL-R-specific variants, we transfected 293 cells with the three constructs expressing sTRAIL, sTRAILDR5 and sTRAILDR4, respectively, harvested the supernatant media after 48 h, adjusted the sTRAIL concentration to 2 ng/ml sTRAIL and transferred the supernatants onto an array of cancer cells. We found cancer cell-specific differences with regard to their propensity to undergo apoptosis via TRAIL-R2 or TRAIL-R1. On one hand, human ovarian cancer A2780 cells, human pancreatic BxPC-3 cells, human colorectal cancer cells HCT116 and Colo205 cells showed increased apoptosis when treated with supernatants from sTRAILDR5-producing 293 cells (Figure 5a), and on the other hand, human pancreatic carcinoma Colo357 cells and human cervical cancer Hela cells exhibited elevated cell death rates with sTRAILDR4 (Figure 5b). The differences in specific apoptosis between the sTRAILDR5 and sTRAILDR4 variants were 38.5 percentage points for A2780 cells, 17.8 percentage points for BxPC-3 cells and 11.5 percentage points and 21.4 percentage points for HCT116 cells and Colo205 cells, respectively. For the cells that predominantly used the TRAIL-R1 pathway, the differences were 7.4 percentage points and 8 percentage points for Colo357 and Hela cells, respectively, in favor of sTRAILDR4. Although these analyses focused on differences between sTRAILDR5 and sTRAILDR4, they did not fully reveal the advantages of the respective TRAIL variants over wild-type sTRAIL. We hypothesized that decreasing sTRAIL concentrations would better unearth such differences. Therefore, we treated Colo205 cells with a lower dose of sTRAIL and its variants (0.5 ng/ml). We found that at this concentration sTRAILDR5 induced apoptosis levels that were equal to those obtained with 2 ng/ml, that is, >95%, whereas apoptosis induction with sTRAIL and sTRAILDR4 dropped to levels measured with control supernatants (Figure 5c). These results demonstrate that sTRAIL variants can have a markedly augmented therapeutic potential as compared with wild-type sTRAIL. Thus, we sought to combine TRAIL-R-specific variants with the MSC-based delivery system.
Apoptosis analyses of a panel of cancer cells reveal divergent responses to sTRAILDR5 and sTRAILDR4. (a) Supernatants from 293 cells that were transfected with enhanced green fluorescent protein (EGFP; gray), sTRAILDR5 (green) and sTRAILDR4 (red) constructs were diluted down to 2 ng/ml TRAIL and then transferred onto A2780, BxPC-3, HCT116 and Colo205 cells. Subsequent apoptosis measurements (24 h) showed that these tumor cells were significantly more responsive to sTRAILDR5 than to sTRAILDR4. Data represent averages±S.E.M. (n=3, P<0.001, P=0.001, P=0.008, P=0.02). (b) Colo357 and Hela cells were incubated with the same EGFP (gray), sTRAILDR5 (green) and sTRAILDR4 (red) 293 cell supernatants (normalized to 2 ng/ml of TRAIL) for 24 h before apoptosis was measured revealing that these tumor cells underwent apoptosis preferentially via TRAIL-R1. Data represent averages±S.E.M. (n=3, P=0.001, P=0.04). (c) Colo205 cells were treated with 293 cell supernatants normalized to a lower TRAIL concentration of 0.5 ng/ml. The apoptosis results for EGFP (gray), sTRAIL (black), sTRAILDR5 (green) and sTRAILDR4 (red) demonstrate that sTRAILDR5 has superior activity not only over sTRAILDR4 but also over sTRAIL at this low concentration. Data represent averages±S.E.M. (n=3, P<0.001, P<0.001)
The sTRAIL variants expressed and secreted from MSCs induce apoptosis in cancer cells
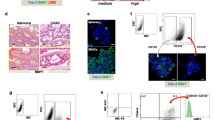
Having established functionality and specificity of the sTRAIL variants, we generated adenoviral vectors with expression cassettes containing the TRAIL-R-specific sTRAIL forms. These adenoviral vectors, termed Ad.sTRAILDR4 and Ad.sTRAILDR5, respectively, were, together with Ad.sTRAIL, used to transduce murine MSCs. The resulting MSCs expressing the sTRAIL variants were labeled MSC.sTRAILDR4 and MSC.sTRAILDR5, respectively. The levels of sTRAIL were visualized by western blot and found to be equal for all three sTRAIL forms (Figure 6a). Mixing MSC.sTRAILDR5 with Colo205 cells at cellular ratios of 1 : 5, 1 : 10 and 1 : 20 led to significantly more apoptosis than with MSC.sTRAIL or MSC.sTRAILDR4, demonstrating the higher susceptibility of Colo205 cells to undergo apoptosis via a TRAIL-R2-mediated pathway (Figure 6b). When we assayed apoptosis at the molecular level, we found that MSC.sTRAILDR5 more profoundly induced caspase-8 activation than MSC.sTRAIL and MSC.sTRAILDR4 (Figure 6b). These results were supported by caspase-3 western blots that showed full caspase-3 activation in MSC.sTRAILDR5-treated Colo205 cells, significantly less in MSC.sTRAIL-stimulated cells and no caspase-3 activation in the MSC.sTRAILDR4 sample (Figure 6b). In contrast, in Hela cells, addition of MSC.sTRAILDR4 gave rise to markedly higher apoptosis levels compared with MSC.sTRAIL and MSC.sTRAILDR5 (Figure 6c). Analyzing caspase-8 activation by western blot revealed that in accordance with the apoptosis results, MSC.sTRAILDR4 gave rise to the strongest caspase-8 activation (Figure 6c). In addition, caspase-3 immunoblots showed that only MSC.sTRAILDR4 caused full activation as evident by the appearance of the active fragments (Figure 6c). Taken together, these results show that MSCs can be utilized to secrete sTRAIL variants that in turn can more effectively induce apoptosis in different cancer cells.
MSC.sTRAILDR5 and MSC.sTRAILDR4 induce increased apoptosis in different cancer cells. (a) Equal TRAIL expression in MSC.sTRAIL, MSC.sTRAILDR5 and MSC.sTRAILDR4 was demonstrated by western blot. (b) MSC.DsRed (gray), MSC.sTRAIL (black), MSC.sTRAILDR5 (green) and MSC.sTRAILDR4 (red) were mixed with Colo205 cells at different ratios (1 : 5, 1 : 10 and 1 : 20) for 24 h. Then, cells were harvested and apoptosis was measured (left). Data represent averages±S.E.M. (n=3, P<0.001). Caspase-8 (right, upper panel) and caspase-3 (right, lower panel) activation of the highest mix ratio (1 : 5) was determined by western blotting analysis. Pro-caspase bands are marked by an (*), whereas cleavage products indicative of activation are indicated by arrows. The CuZn superoxide dismutase (CuZnSOD) blot served as loading control. (c) MSC.DsRed (gray), MSC.sTRAIL (black), MSC.sTRAILDR5 (green) and MSC.sTRAILDR4 (red) were mixed with Hela cells at different ratios (1 : 5, 1 : 10 and 1 : 20) for 24 h before apoptosis was measured (left). Data represent averages±S.E.M. (n=3, P=0.002, P=0.002). Caspase-8 (right, upper panel) and caspase-3 (right, lower panel) western blots show the different caspase-8 and caspase-3 activation levels in an experiment, in which MSCs and Hela cells were mixed 1 : 5. Pro-caspase-8 and −3 are marked by an (*) and proteolytic cleavage fragments are indicated by arrows. The CuZnSOD blot is the loading control
Combination treatments enhance the effects of MSC.sTRAILDR5 and MSC.sTRAILDR4 in colon and pancreatic cancer cells
After having demonstrated the utility of 5-FU sensitization to MSC.sTRAIL-induced apoptosis in vivo on one side as well as the potential of TRAIL-R-specific variants on the other, we sought to combine these two approaches. In particular, as we had found that 5-FU sensitization to TRAIL was mediated by TRAIL-R2 and its upregulation, we hypothesized that TRAIL-R2-specific variants could afford superior cancer cell killing effects in this context. We pretreated HCT116 cells with 5-FU for 24 h, after which MSC.sTRAIL, MSC.sTRAILDR5 and MSC.sTRAILDR4 were added for another 24 h, before apoptosis was measured. We found that pretreatment with 5-FU led to significantly increased apoptosis levels after treatment with MSC.sTRAILDR5 compared with MSC.sTRAIL and MSC.sTRAILDR4 (Figure 7a). To study the specificity of MSC.sTRAILDR4 and MSC.sTRAILDR5 in combination with 5-FU, we mixed them with HCT116, HCT.shDR4 and HCT.shDR5 cells that had been pretreated with 5-FU. Our results show that in pretreated HCT.shDR4 and HCT116 cells, MSC.sTRAILDR5 showed higher apoptosis levels whereas in HCT.shDR5 the 5-FU sensitization effect was blocked, demonstrating that it was mediated by TRAIL-R2 (Figure 7a). Noteworthy, MSC.sTRAILDR4 exerted augmented apoptosis-inducing activity on HCT.shDR5, as no TRAIL-R2 could impede the formation of TRAIL-R1 homotrimers, resulting in more efficient apoptosis induction by the TRAIL-R1-specific variant (Figure 7a). Accordingly, MSC.sTRAILDR5 gave rise to more cell death in 5-FU-pretreated HCT.shDR4 cells (Figure 7a). Thus, MSC.sTRAILDR5 is the preferred system in combination with 5-FU in this case. However, in addition to Hela and Colo357 cells (Figure 5b), we and others have shown that in pancreatic cancer cells TRAIL-induced apoptosis is preferentially mediated via TRAIL-R1 and is profoundly enhanced by XIAP inhibition.29, 30 Therefore, to also test utility of TRAIL-R1-specific variants in conjunction with an alternative sensitization approach to 5-FU, namely XIAP inhibition, we generated stable XIAP knockdown clones of the TRAIL-resistant pancreatic PancTu1 cancer cell line. These clones were named PancTu1.shXIAP and were shown to harbor no detectable XIAP levels by western blot (Figure 7b). When we treated PancTu1.shXIAP cells with MSC.sTRAIL, MSC.sTRAILDR4 and MSC.sTRAILDR5, we could measure apoptosis in all three cases, but MSC.sTRAILDR4 gave rise to significantly higher cell death rates than MSC.sTRAIL and MSC.sTRAILDR5 (Figure 7b). These findings are in line with our previous findings29, 30 and underline that pancreatic cancer cells undergo apoptosis preferentially via TRAIL-R1. In summary, TRAIL-R-specific variants offer a substantial improvement over normal TRAIL as they can better target TRAIL-resistant cancer cells and act more specifically and effectively in combined treatment approaches.
Cancer cells can be sensitized to treatment with MSC.sTRAIL variants by silencing of XIAP and 5-FU pretreatment. (a) HCT116 cells were pretreated with 10 μM 5-FU for 24 h before they were mixed with MSC.DsRed (gray), MSC.sTRAIL (black), MSC.sTRAILDR5 (green) and MSC.sTRAILDR4 (red) at different ratios (1 : 5, 1 : 10 and 1 : 20) for another 24 h before apoptosis was measured. The apoptosis rate with MSC.sTRAILDR5 was significantly increased after pretreatment with 5-FU (left). Data represent averages±S.E.M. (n=3, P=0.007, P=0.05, P=0.01). In order to demonstrate specificity, HCT.shDR5 (center) and HCT.shDR4 (right) were also mixed with MSC.DsRed (gray), MSC.sTRAIL (black), MSC.sTRAILDR5 (green) and MSC.sTRAILDR4 (red) at different ratios (1 : 5, 1 : 10 and 1 : 20) and apoptosis was measured 24 h later. Data represent averages±S.E.M. (n=3, P=0.006, P<0.001; n=3, P<0.001, P<0.001, P<0.001). (b) XIAP western blot from protein lysates of PancTu1.shctrl and PancTu1.shXIAP cells. The CuZn superoxide dismutase (CuZnSOD) blot served as loading control (left). Mixing of MSC.DsRed (gray), MSC.sTRAIL (black), MSC.sTRAILDR5 (green) and MSC.sTRAILDR4 (red) at different ratios (1 : 5, 1 : 10 and 1 : 20) with PancTu1.shXIAP cells for 48 h. Subsequent apoptosis measurements showed that in PancTu1.shXIAP cells MSC.sTRAILDR4 had the best effect (right). Data represent averages±S.E.M. (n=3, P<0.001, P=0.02, P<0.001)
Discussion
MSCs are pluripotent adult stem cells that are normally derived from bone marrow and have the potential to differentiate in vitro along numerous mesenchymal lineages.31 As MSCs have been shown to infiltrate tumor tissues,21, 22 they have been employed in various experimental cancer therapeutic approaches, including those that make use of their proposed intrinsic anticancer properties or as a delivery vehicle for therapeutic anticancer transgenes.32 To this end, we and others have shown that MSCs expressing TRAIL led to growth retardation or remission of human lung, glioblastoma, breast and pancreatic carcinoma xenografts, respectively, as well as inhibition of metastasis formation.17, 19, 33, 34, 35 In order to increase the effects of MSC-mediated TRAIL delivery further, we analyzed pretreatment of cancer cells with 5-FU in combination with MSC.sTRAIL. 5-FU is a pyrimidine analog that is used in the treatment of cancer.36 It belongs to the family of drugs called antimetabolites and works through irreversible inhibition of thymidylate synthase. 5-FU has been shown to sensitize to TRAIL-induced apoptosis via various mechanisms including upregulation of TRAIL-R expression and/or downregulation of the anti-apoptotic protein cFlip.26 As its principal use is in colorectal cancer, in which it has been the established form of chemotherapy for decades, we tested 5-FU pretreatment followed by MSC.sTRAIL in HCT116 human colorectal cancer cells. We found pretreating with low doses of 5-FU greatly enhanced apoptosis when the HCT116 cells were subsequently mixed with MSC.sTRAIL. Furthermore, when we established HCT116 xenografts in mice, pretreated them with 5-FU followed by systemic administration of MSC.sTRAIL, we saw complete remission of the tumors as compared with partial responses in the cohort treated with MSC.sTRAIL alone. HCT116 cells are p53 proficient, whereas a substantial fraction of tumors are p53 mutated and/or dysfunctional. Therefore, in order to assess whether this sensitization strategy would also be applicable in p53-defective tumor cells, we analyzed p53-mutated and p53-negative colon cancer cells, HT-29 and HCTp53−/− cells, respectively. We were able to sensitize both cell lines to the effects of TRAIL by 5-FU pretreatment, demonstrating that the approach also has utility in p53-deficient tumors. Subsequently, we sought to elucidate the underlying molecular mechanism of the 5-FU sensitization in order to be able to design further improvements into the MSC.sTRAIL system. We found that this sensitization effect was caused by 5-FU-induced, p53-independent upregulation of TRAIL-R2, as RNAi-mediated silencing of TRAIL-R2, but not of TRAIL-R1, resulted in inhibition of 5-FU/TRAIL-induced apoptosis. Hence, we set out to develop MSCs that express and secrete TRAIL variants that specifically target TRAIL-R2 to achieve greater apoptosis induction in 5-FU-treated tumor cells. First, we cloned the cDNAs coding for TRAIL variants that acted through TRAIL-R1, as control, and TRAIL-R2, respectively, into adenoviral expression constructs, which were used to transduce MSCs, giving rise to MSC.sTRAILDR4 and MSC.sTRAILDR5. Indeed, when we mixed 5-FU-stimulated HCT116 cells with MSC.sTRAIL, MSC.sTRAILDR4 or MSC.sTRAILDR5, we measured the highest apoptosis levels in the cancer cells that were treated with MSC.sTRAILDR5. Taken together, a targeted therapy with MSC.sTRAILDR5 in combination with 5-FU appears to be a viable treatment approach even for p53-deficient tumors.
However, many tumors are resistant to TRAIL-induced apoptosis owing to mechanisms downstream of the receptor and the DISC, which means that 5-FU-mediated TRAIL-R upregulation (alone) would not be a viable sensitization strategy. To this end, we and others have documented the importance of the antiapoptotic protein XIAP, known to be frequently upregulated in cancers, and its utility as a therapeutic target.37, 38 Pancreatic cancers in particular are known to be aggressive and resistant to apoptosis at least in part owing to high XIAP levels.39 In the present study, we generated stable XIAP knockdown clones by RNAi in the pancreatic cancer cell line PancTu1 and treated them with our array of MSC.sTRAIL variants. We found that in contrast to their parental counterparts, XIAP-silenced cells were sensitive to the effects of MSC-produced sTRAIL,19 and also found that in this context PancTu1 responded better to TRAIL-R1-specific sTRAIL, as we have reported previously in a similar approach using agonistic TRAIL-R1-specific antibodies.30 As several XIAP/IAP inhibitors are currently in clinical tests, the silencing of XIAP or its functions is a potential approach and can be achieved either pharmacologically or through improved RNAi tools40, 41, 42 in a potential, future clinical setting. Furthermore, although MSC.sTRAILDR5 were the optimal system in particular for 5-FU-primed colorectal cancer cells, the effects on other tumor cells that were not treated with 5-FU were more divergent. In Colo205, HCT116, BxPc-3 and A2780 cells, sTRAILDR5 showed the best effects, whereas sTRAILDR4 induced more apoptosis in Hela, Colo357 and PancTu1.shXIAP cells, indicating that in certain cases TRAIL-R1-specific variants are more efficacious. Therefore, it would be valuable to be able to predict the responsiveness of cancer cells to the different TRAIL variants. To this end, we have previously reported that the expression levels of the different TRAIL-Rs can be a predicting factor for TRAIL responsiveness.43 In line with those results, on Colo205 cells, which trigger apoptosis preferentially via TRAIL-R2, TRAIL-R1 is undetectable. Thus, although the predictive potential of TRAIL-R expression is limited, it can still serve as a first-line test aiding in deciding which TRAIL variant to use. Overall, our results demonstrate that TRAIL-R-specific TRAIL variants are significantly more efficacious than wild-type TRAIL, in particular in combination with 5-FU.
Materials and Methods
Reagents and cell culture
All chemicals and reagents unless otherwise stated, were purchased from Sigma (St Louis, MO, USA). Media were purchased from Invitrogen (Carlsbad, CA, USA) unless otherwise stated. Recombinant TRAIL was from R&D Systems (Minneapolis, MN, USA) and zVAD from Santa Cruz (Santa Cruz, CA, USA). Antibodies and cells are described in the Supplementary Materials and Methods section. 293 cells were transiently transfected using the calcium phosphate transfection method, which is described in more detail in the Supplementary Materials and Methods section.
Apoptosis assay
Apoptosis was measured according to Nicoletti et al.44 and has been described previously.45 Please see Supplementary Materials and Methods for detail.
Generation of adenoviral vectors and transduction of MSCs
Recombinant E1/E3-deleted adenoviral vectors (Ad.sTRAIL, Ad.sTRAILDR4, Ad.sTRAILDR5 and control Ad.DsRed) were generated using the ViraPower adenoviral expression system (Invitrogen) as described earlier.34 Transduction of MSCs was carried out as described earlier.34 TRAIL variants, sTRAILDR5 (D269H) and sTRAILDR4 (S159R), were generated using the Quick-Change site-directed mutagenesis kit (Stratagene, La Jolla, CA, USA) and confirmed by DNA sequencing. Please see Supplementary Materials and Methods for detail.
Isolation, expansion and differentiation assays of MSCs
MSCs from 4- to 6-week old FVB/N (FVB/n mouse strain) mice were prepared based on the method of Peister et al.46 The cells were grown in Dulbecco’s modified Eagle’s medium (DMEM)–low glucose with 100 U/ml penicillin, 100 μg/ml streptomycin and 15% fetal bovine serum (FBS). Differentiation assays for MSCs were carried out as described previously.21 The murine MSCs (mMSCs) are generally referred to as MSCs throughout the text.
Cell surface staining and FACS analyses
The surface expression of TRAIL-Rs was measured by incubating cells with R-phycoerythrin (PE)-conjugated mouse anti-human TRAIL-R1 and mouse anti-human TRAIL-R2 as described previously.34 The same procedure was used to stain for CD11b, CD19, CD117, CD45, TER-119, CD29, Sca-1 and CD44.19
Mixing of MSCs with tumor cells
Tumor cells were plated in six-well plates at 105 cells per well and left overnight to adhere. Then, cells were pretreated with 10 μM 5-FU or left untreated. Adenovirus-transduced MSCs (48 h) were then added 24 h later at different ratios in fresh medium. Another 24 h later, cells were collected for apoptosis assays. The supernatants of adenovirally transduced MSCs were diluted (1 : 5) and also added onto HCT116 cells.
Animal studies
Ten-week-old female CD1 nu/nu (Crl:CD1-Foxn1nu) mice (Charles River, Wilmington, MA, USA) were injected with 5 × 106 HCT116 cells in 200 μl phosphate-buffered saline (PBS). After 10 days, and when tumors were palpable, the animals were intraperitoneally injected with 5-FU (150 mg/kg) and intravenously with 1 × 105 MSCs that had been transduced with the respective adenoviral vectors 24 h earlier. The injections were repeated 10 days later. The growth of the tumors was then followed over 33 days. The tumor volume was calculated using three different diameters and the formula: π/6 × (d1 × d2 × d3). The animal studies were performed according to national laws and covered by license from the Irish government.
Histological analyses
Please see Supplementary Materials and Methods for detail.
Western blot
Western blots were principally carried out as described previously.47 Please see Supplementary Materials and Methods for detail.
TRAIL enzyme-linked immunosorbent assay (ELISA)
To examine secreted sTRAIL, the culture supernatants of transfected 293 cells and transduced MSCs were cleared by centrifugation at 2000 × g for 20 min. The supernatants were diluted and a commercial TRAIL ELISA kit (R&D Systems) was used following the manufacturer’s instructions. Please see Supplementary Materials and Methods for detail.
RNAi knockdown constructs and stable cell line generation
Small hairpin (sh) RNA motifs were expressed inside pU6.ENTR and pBlock-iT plasmids (Invitrogen). Please see Supplementary Materials and Methods for sequence information and further details.
Statistical analysis
If not stated, three independent experiments were performed. Experimental values are expressed as mean value±S.E.M. For significance analyses, analysis of variance (ANOVA) between groups was used and *P≤0.05 was considered significant, **P≤0.01 as highly significant and ***P≤0.001 as extremely significant.
Abbreviations
- 5-FU:
-
5-fluorouracil
- Ad:
-
adenoviral
- ANOVA:
-
analysis of variance
- CD1 nu/nu:
-
Crl:CD1-Foxn1nu
- cFlip:
-
cellular FLICE-like inhibitory protein
- CMV:
-
cytomegalovirus
- DcR1:
-
decoy receptor 1
- DcR2:
-
decoy receptor 2
- DD:
-
death domain
- DISC:
-
death inducing signaling complex
- DMEM:
-
Dulbecco’s modified Eagle’s medium
- DR4:
-
death receptor 4
- DR5:
-
death receptor 5
- DsRed:
-
Discosoma sp. red fluorescent protein
- ELISA:
-
enzyme-linked immunosorbent assay
- FACS:
-
fluorescence-activated cell sorting
- FADD:
-
Fas-associated protein with death domain
- FBS:
-
fetal bovine serum
- hFIB:
-
human Fibrillin-1 gene
- FVB/N:
-
FVB/n mouse strain
- Furin CS:
-
furin cleavage site
- H&E:
-
hematoxylin and eosin
- IAP:
-
inhibitor of apoptosis
- ILZ:
-
isoleucine zipper
- MSC:
-
mesenchymal stem cell
- mMSC:
-
murine MSC
- OPG:
-
osteoprotegerin
- p53:
-
protein 53
- PBS:
-
phosphate-buffered saline
- PCNA:
-
proliferating cell nuclear antigen
- PE:
-
R-phycoerythrin
- RNAi:
-
RNA interference
- TNF:
-
tumor necrosis factor
- TRAIL:
-
TNF-related apoptosis-inducing ligand
- rTRAIL:
-
recombinant TRAIL
- sTRAIL:
-
soluble TRAIL
- TRAIL-R:
-
TRAIL-receptor
- XIAP:
-
X-linked inhibitor of apoptosis protein
- zVAD:
-
carbobenzoxy-valyl-alanyl-aspartyl-[O-methyl]-fluoromethylketone
References
Papenfuss K, Cordier SM, Walczak H . Death receptors as targets for anti-cancer therapy. J Cell Mol Med 2008; 12: 2566–2585.
Zhang L, Ren X, Alt E, Bai X, Huang S, Xu Z et al. Chemoprevention of colorectal cancer by targeting APC-deficient cells for apoptosis. Nature 2010; 464: 1058–1061.
Johnstone RW, Frew AJ, Smyth MJ . The TRAIL apoptotic pathway in cancer onset, progression and therapy. Nat Rev Cancer 2008; 8: 782–798.
Sprick MR, Weigand MA, Rieser E, Rauch CT, Juo P, Blenis J et al. FADD/MORT1 and caspase-8 are recruited to TRAIL receptors 1 and 2 and are essential for apoptosis mediated by TRAIL receptor 2. Immunity 2000; 12: 599–609.
Bratton SB, MacFarlane M, Cain K, Cohen GM . Protein complexes activate distinct caspase cascades in death receptor and stress-induced apoptosis. Exp Cell Res 2000; 256: 27–33.
Camidge DR, Herbst RS, Gordon MS, Eckhardt SG, Kurzrock R, Durbin B et al. A phase I safety and pharmacokinetic study of the death receptor 5 agonistic antibody PRO95780 in patients with advanced malignancies. Clin Cancer Res 2010; 16: 1256–1263.
Chuntharapai A, Dodge K, Grimmer K, Schroeder K, Marsters SA, Koeppen H et al. Isotype-dependent inhibition of tumor growth in vivo by monoclonal antibodies to death receptor 4. J Immunol 2001; 166: 4891–4898.
Ichikawa K, Liu W, Zhao L, Wang Z, Liu D, Ohtsuka T et al. Tumoricidal activity of a novel anti-human DR5 monoclonal antibody without hepatocyte cytotoxicity. Nat Med 2001; 7: 954–960.
Trarbach T, Moehler M, Heinemann V, Kohne CH, Przyborek M, Schulz C et al. Phase II trial of mapatumumab, a fully human agonistic monoclonal antibody that targets and activates the tumour necrosis factor apoptosis-inducing ligand receptor-1 (TRAIL-R1), in patients with refractory colorectal cancer. Br J Cancer 2010; 102: 506–512.
MacFarlane M, Kohlhaas SL, Sutcliffe MJ, Dyer MJ, Cohen GM . TRAIL receptor-selective mutants signal to apoptosis via TRAIL-R1 in primary lymphoid malignancies. Cancer Res 2005; 65: 11265–11270.
Reis CR, van der Sloot AM, Natoni A, Szegezdi E, Setroikromo R, Meijer M et al. Rapid and efficient cancer cell killing mediated by high-affinity death receptor homotrimerizing TRAIL variants. Cell Death Dis 2010; 1: e83.
Duiker EW, de Vries EG, Mahalingam D, Meersma GJ, Boersma-van Ek W, Hollema H et al. Enhanced antitumor efficacy of a DR5-specific TRAIL variant over recombinant human TRAIL in a bioluminescent ovarian cancer xenograft model. Clin Cancer Res 2009; 15: 2048–2057.
van der Sloot AM, Tur V, Szegezdi E, Mullally MM, Cool RH, Samali A et al. Designed tumor necrosis factor-related apoptosis-inducing ligand variants initiating apoptosis exclusively via the DR5 receptor. Proc Natl Acad Sci USA 2006; 103: 8634–8639.
Ashkenazi A, Holland P, Eckhardt SG . Ligand-based targeting of apoptosis in cancer: the potential of recombinant human apoptosis ligand 2/Tumor necrosis factor-related apoptosis-inducing ligand (rhApo2L/TRAIL). J Clin Oncol 2008; 26: 3621–3630.
Ashkenazi A, Pai RC, Fong S, Leung S, Lawrence DA, Marsters SA et al. Safety and antitumor activity of recombinant soluble Apo2 ligand. J Clin Invest 1999; 104: 155–162.
Griffith TS, Stokes B, Kucaba TA, Earel JK, VanOosten RL, Brincks EL et al. TRAIL gene therapy: from preclinical development to clinical application. Curr Gene Ther 2009; 9: 9–19.
Kim SM, Lim JY, Park SI, Jeong CH, Oh JH, Jeong M et al. Gene therapy using TRAIL-secreting human umbilical cord blood-derived mesenchymal stem cells against intracranial glioma. Cancer Res 2008; 68: 9614–9623.
Loebinger MR, Eddaoudi A, Davies D, Janes SM . Mesenchymal stem cell delivery of TRAIL can eliminate metastatic cancer. Cancer Res 2009; 69: 4134–4142.
Mohr A, Albarenque SM, Deedigan L, Yu R, Reidy M, Fulda S et al. Targeting of XIAP combined with systemic mesenchymal stem cell-mediated delivery of sTRAIL ligand inhibits metastatic growth of pancreatic carcinoma cells. Stem Cells 2010; 28: 2109–2120.
Mohr A, Henderson G, Dudus L, Herr I, Kuerschner T, Debatin KM et al. AAV-encoded expression of TRAIL in experimental human colorectal cancer leads to tumor regression. Gene Ther 2004; 11: 534–543.
Albarenque SM, Zwacka RM, Mohr A . Both human and mouse mesenchymal stem cells promote breast cancer metastasis. Stem Cell Res 2011; 7: 163–171.
Hall B, Dembinski J, Sasser AK, Studeny M, Andreeff M, Marini F . Mesenchymal stem cells in cancer: tumor-associated fibroblasts and cell-based delivery vehicles. Int J Hematol 2007; 86: 8–16.
Luetzkendorf J, Mueller LP, Mueller T, Caysa H, Nerger K, Schmoll HJ . Growth-inhibition of colorectal carcinoma by lentiviral TRAIL-transgenic human mesenchymal stem cells requires their substantial intratumoral presence. J Cell Mol Med 2009; 14: 2292–2304.
Wu GS . TRAIL as a target in anti-cancer therapy. Cancer Lett 2009; 285: 1–5.
Kim MH, Billiar TR, Seol DW . The secretable form of trimeric TRAIL, a potent inducer of apoptosis. Biochem Biophys Res Commun 2004; 321: 930–935.
Ganten TM, Haas TL, Sykora J, Stahl H, Sprick MR, Fas SC et al. Enhanced caspase-8 recruitment to and activation at the DISC is critical for sensitisation of human hepatocellular carcinoma cells to TRAIL-induced apoptosis by chemotherapeutic drugs. Cell Death Differ 2004; 11 (Suppl 1): S86–S96.
Wu GS, Burns TF, McDonald ER, Jiang W, Meng R, Krantz ID et al. KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet 1997; 17: 141–143.
Schymkowitz JW, Rousseau F, Martins IC, Ferkinghoff-Borg J, Stricher F, Serrano L . Prediction of water and metal binding sites and their affinities by using the Fold-X force field. Proc Natl Acad Sci USA 2005; 102: 10147–10152.
Lemke J, Noack A, Adam D, Tchikov V, Bertsch U, Roder C et al. TRAIL signaling is mediated by DR4 in pancreatic tumor cells despite the expression of functional DR5. J Mol Med 2010; 88: 729–740.
Stadel D, Mohr A, Ref C, MacFarlane M, Zhou S, Humphreys R et al. TRAIL-induced apoptosis is preferentially mediated via TRAIL receptor 1 in pancreatic carcinoma cells and profoundly enhanced by XIAP inhibitors. Clin Cancer Res 2010; 16: 5734–5749.
Barry FP, Murphy JM . Mesenchymal stem cells: clinical applications and biological characterization. Int J Biochem Cell Biol 2004; 36: 568–584.
Nakamizo A, Marini F, Amano T, Khan A, Studeny M, Gumin J et al. Human bone marrow-derived mesenchymal stem cells in the treatment of gliomas. Cancer Res 2005; 65: 3307–3318.
Menon LG, Kelly K, Yang HW, Kim SK, Black PM, Carroll RS . Human bone marrow-derived mesenchymal stromal cells expressing S-TRAIL as a cellular delivery vehicle for human glioma therapy. Stem Cells 2009; 27: 2320–2330.
Mohr A, Lyons M, Deedigan L, Harte T, Shaw G, Howard L et al. Mesenchymal stem cells expressing TRAIL lead to tumour growth inhibition in an experimental lung cancer model. J Cell Mol Med 2008; 12: 2628–2643.
Sasportas LS, Kasmieh R, Wakimoto H, Hingtgen S, van de Water JA, Mohapatra G et al. Assessment of therapeutic efficacy and fate of engineered human mesenchymal stem cells for cancer therapy. Proc Natl Acad Sci USA 2009; 106: 4822–4827.
Sampath D, Rao VA, Plunkett W . Mechanisms of apoptosis induction by nucleoside analogs. Oncogene 2003; 22: 9063–9074.
Braeuer SJ, Buneker C, Mohr A, Zwacka RM . Constitutively activated nuclear factor-kappaB, but not induced NF-kappaB, leads to TRAIL resistance by up-regulation of X-linked inhibitor of apoptosis protein in human cancer cells. Mol Cancer Res 2006; 4: 715–728.
Vogler M, Durr K, Jovanovic M, Debatin KM, Fulda S . Regulation of TRAIL-induced apoptosis by XIAP in pancreatic carcinoma cells. Oncogene 2007; 26: 248–257.
Vogler M, Walczak H, Stadel D, Haas TL, Genze F, Jovanovic M et al. Targeting XIAP bypasses Bcl-2-mediated resistance to TRAIL and cooperates with TRAIL to suppress pancreatic cancer growth in vitro and in vivo. Cancer Res 2008; 68: 7956–7965.
Davis ME . The first targeted delivery of siRNA in humans via a self-assembling, cyclodextrin polymer-based nanoparticle: from concept to clinic. Mol Pharmacol 2009; 6: 659–668.
Oost TK, Sun C, Armstrong RC, Al Assaad AS, Betz SF, Deckwerth TL et al. Discovery of potent antagonists of the antiapoptotic protein XIAP for the treatment of cancer. J Med Chem 2004; 47: 4417–4426.
Vogler M, Walczak H, Stadel D, Haas TL, Genze F, Jovanovic M et al. Small molecule XIAP inhibitors enhance TRAIL-induced apoptosis and antitumor activity in preclinical models of pancreatic carcinoma. Cancer Res 2009; 69: 2425–2434.
Buneker C, Mohr A, Zwacka RM . The TRAIL-receptor-1: TRAIL-receptor-3 and -4 ratio is a predictor for TRAIL sensitivity of cancer cells. Oncol Rep 2009; 21: 1289–1295.
Nicoletti I, Migliorati G, Pagliacci MC, Grignani F, Riccardi C . A rapid and simple method for measuring thymocyte apoptosis by propidium iodide staining and flow cytometry. J Immunol Methods 1991; 139: 271–279.
Mohr A, Buneker C, Gough RP, Zwacka RM . MnSOD protects colorectal cancer cells from TRAIL-induced apoptosis by inhibition of Smac/DIABLO release. Oncogene 2008; 27: 763–774.
Peister A, Mellad JA, Larson BL, Hall BM, Gibson LF, Prockop DJ . Adult stem cells from bone marrow (MSCs) isolated from different strains of inbred mice vary in surface epitopes, rates of proliferation, and differentiation potential. Blood 2004; 103: 1662–1668.
Zwacka RM, Stark L, Dunlop MG . NF-kappaB kinetics predetermine TNF-alpha sensitivity of colorectal cancer cells. J Gene Med 2000; 2: 334–343.
Acknowledgements
We thank the members of the TRIDENT consortium for helpful discussions and input and are grateful to Chirlei Klein Büneker, Jérôme Courtête and Mairéad Reidy for their assistance during the initial stages of the project. The work was supported by an EU-FP6-STREP (TRIDENT) award. The work was also supported by an EU-FP6 Marie-Curie Excellence Team Award (MIST), by an EU-RTN Award (ApopTrain) and a Research Frontier Project Grant from Science Foundation Ireland (BIM084) (to RMZ).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Edited by A Stephanou
Supplementary Information accompanies the paper on Cell Death and Disease website
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Yu, R., Deedigan, L., Albarenque, S. et al. Delivery of sTRAIL variants by MSCs in combination with cytotoxic drug treatment leads to p53-independent enhanced antitumor effects. Cell Death Dis 4, e503 (2013). https://doi.org/10.1038/cddis.2013.19
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2013.19
Keywords
This article is cited by
-
Generation of magnetic biohybrid microrobots based on MSC.sTRAIL for targeted stem cell delivery and treatment of cancer
Cancer Nanotechnology (2023)
-
Mesenchymal stem cells in cancer progression and anticancer therapeutic resistance
Cancer Cell International (2021)
-
TRAIL-receptor preferences in pancreatic cancer cells revisited: Both TRAIL-R1 and TRAIL-R2 have a licence to kill
BMC Cancer (2015)
-
c-Rel is a critical mediator of NF-κB-dependent TRAIL resistance of pancreatic cancer cells
Cell Death & Disease (2014)
-
Claudin-7 suppresses the cytotoxicity of TRAIL-expressing mesenchymal stem cells in H460 human non-small cell lung cancer cells
Apoptosis (2014)