Article
|
Open Access
Featured
-
-
Article
| Open Access Signal transduction controls heterogeneous NF-κB dynamics and target gene expression through cytokine-specific refractory states
Signal transduction controls heterogeneous NF-κB dynamics and target gene expression through cytokine-specific refractory states
In biological systems, timing is often critical to the interpretation of signals that determine cell fate. Here the authors demonstrate how single cells and cellular populations respond dynamically to pulsatile stimulation by TNFα and IL-1β, and suggest a mechanism by which the two cytokines can synergistically modulate inflammation.
- Antony Adamson
- , Christopher Boddington
- & Pawel Paszek
-
Article
| Open Access Genomics-informed isolation and characterization of a symbiotic Nanoarchaeota system from a terrestrial geothermal environment
Genomics-informed isolation and characterization of a symbiotic Nanoarchaeota system from a terrestrial geothermal environment
Many microbial lineages have not yet been cultured, which hampers our understanding of their physiology. Here, Wurch et al. use single-cell genomics to infer cultivation conditions for the isolation of a tiny ectosymbiotic nanoarchaeon and its crenarchaeota host from a geothermal spring.
- Louie Wurch
- , Richard J. Giannone
- & Mircea Podar
-
Article
| Open Access Targeted proteomics identifies liquid-biopsy signatures for extracapsular prostate cancer
Targeted proteomics identifies liquid-biopsy signatures for extracapsular prostate cancer
Proteomic technologies are capable of identifying thousands of proteins in biological samples, but biomarker applications are lagging. Here the authors use Multiple Reaction Monitoring Mass Spectrometry to delineate peptide signatures that accurately distinguish between defined prostate cancer patient risk groups.
- Yunee Kim
- , Jouhyun Jeon
- & Thomas Kislinger
-
Article
| Open Access A dynamic pathway analysis approach reveals a limiting futile cycle in N-acetylglucosamine overproducing Bacillus subtilis
A dynamic pathway analysis approach reveals a limiting futile cycle in N-acetylglucosamine overproducing Bacillus subtilis
Rate-limiting steps in synthetic metabolic pathways are difficult to identify. Here, the authors monitor metabolite dynamics and apply kinetic modelling during the start-up phase of the Bacillus subtilisGlcNAc pathway to discover a futile cycle, allowing them to identify a more productive strain.
- Yanfeng Liu
- , Hannes Link
- & Uwe Sauer
-
Article
| Open Access Structure and inference in annotated networks
Structure and inference in annotated networks
Analysis of network structure is usually based on knowledge of connections alone, ignoring additional information such as gender or age of individuals in social networks. Here the authors devise an approach that incorporates such metadata and uses it to improve the detection of network communities.
- M. E. J. Newman
- & Aaron Clauset
-
Article
| Open Access The Parkinson’s disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway
The Parkinson’s disease-associated genes ATP13A2 and SYT11 regulate autophagy via a common pathway
Mutations in ATP13A2 are associated with lysosomal dysfunction and early onset Parkinson’s disease. Here Bento et al. show that ATP13A2 depletion negatively regulates SYT11, at both transcriptional and post-translational levels, which in turn impairs function of the autophagy-lysosome pathway.
- Carla F. Bento
- , Avraham Ashkenazi
- & David C. Rubinsztein
-
Article
| Open Access Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow
Improving GENCODE reference gene annotation using a high-stringency proteogenomics workflow
Identifying and annotating functional elements in the human genome remains a challenging but important task. Here the authors propose a priority annotation score to rank identifications and suggest how proteogenomics evidence can be interpreted and what additional information substantiates protein-coding potential for annotation.
- James C. Wright
- , Jonathan Mudge
- & Jennifer Harrow
-
Article
| Open Access The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2)
The dynamic transcriptional and translational landscape of the model antibiotic producer Streptomyces coelicolor A3(2)
Bacteria of the genus Streptomyces produce a great variety of natural products, the biosynthesis of which is subject to complex regulatory networks. Here the authors present a high-resolution, genome-wide analysis of the transcriptome and translatome of Streptomyces coelicolorunder various growth conditions.
- Yujin Jeong
- , Ji-Nu Kim
- & Byung-Kwan Cho
-
Article
| Open Access Geometrical assembly of ultrastable protein templates for nanomaterials
Geometrical assembly of ultrastable protein templates for nanomaterials
Protein nanotechnology for the fabrication of protein-based nanoscale devices is gaining momentum but assembling well-defined three-dimensional shapes is still challenging. Here, the authors use an existing prefoldin assembled system to design a template for the construction of geometrically constrained structures.
- Dominic J. Glover
- , Lars Giger
- & Douglas S. Clark
-
Article
| Open Access Cox process representation and inference for stochastic reaction–diffusion processes
Cox process representation and inference for stochastic reaction–diffusion processes
Stochastic reaction-diffusion systems are used for modelling spatial dynamics in many disciplines, but parameter inference and model selection remain challenging. Here the authors offer a solution enabled by a connection between reaction-diffusion and the well-studied spatio-temporal Cox processes.
- David Schnoerr
- , Ramon Grima
- & Guido Sanguinetti
-
Article
| Open Access The fin-to-limb transition as the re-organization of a Turing pattern
The fin-to-limb transition as the re-organization of a Turing pattern
Mouse digit patterning is controlled by a Turing network of Bmp, Sox9, and Wnt. Here, Onimaru et al. show that fin patterning in the catshark, Scyliorhinus canicula, is controlled by the same network with a different spatial organization; thus, the Turing network is deeply conserved in limb development.
- Koh Onimaru
- , Luciano Marcon
- & James Sharpe
-
Article
| Open Access Adaptive evolution of complex innovations through stepwise metabolic niche expansion
Adaptive evolution of complex innovations through stepwise metabolic niche expansion
A fundamental question in evolutionary biology is how complex innovations requiring multiple genetic changes arise. Here the authors provide lines of evidence that changing environments facilitate the adaptive evolution of complex metabolic innovations via stepwise acquisition of single reactions.
- Balázs Szappanos
- , Jonathan Fritzemeier
- & Balázs Papp
-
Article
| Open Access A microfluidics-based in vitro model of the gastrointestinal human–microbe interface
A microfluidics-based in vitro model of the gastrointestinal human–microbe interface
Research on the interactions between the gut microbiota and human cells would greatly benefit from improved in vitro models. Here, Shah et al. present a modular microfluidics-based model that allows co-culture of human and microbial cells followed by 'omic' molecular analyses of the two cell contingents.
- Pranjul Shah
- , Joëlle V. Fritz
- & Paul Wilmes
-
Article
| Open Access Inhibitory interactions promote frequent bistability among competing bacteria
Inhibitory interactions promote frequent bistability among competing bacteria
We know little about the effect of relationships between species on the assembly of microbial communities. Here the authors map pairwise invasion relations between bacteria and find that instead of one strain dominating, inhibitory interactions mean that often neither strain can invade the other.
- Erik S. Wright
- & Kalin H. Vetsigian
-
Article
| Open Access Whi5 phosphorylation embedded in the G1/S network dynamically controls critical cell size and cell fate
Whi5 phosphorylation embedded in the G1/S network dynamically controls critical cell size and cell fate
In budding yeast the G1/S transition requires the attainment of a critical cell size. Here the authors unravel its basic control mechanism by integrating simulations of a mathematical model of multisite phosphorylation of Whi5 by Cln3–Cdk1, with molecular analyses of a Whi5 phospho-mimetic mutant.
- Pasquale Palumbo
- , Marco Vanoni
- & Lilia Alberghina
-
Article
| Open Access A geometrical approach to control and controllability of nonlinear dynamical networks
A geometrical approach to control and controllability of nonlinear dynamical networks
Complex networks, including physical, biological and social systems are ubiquitous, but understanding of how to control them is elusive. Here Wang et al. develop a framework based on the concept of attractor networks to facilitate the study of controllability of nonlinear dynamics in complex systems.
- Le-Zhi Wang
- , Ri-Qi Su
- & Ying-Cheng Lai
-
Article
| Open Access A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells
A programmable synthetic lineage-control network that differentiates human IPSCs into glucose-sensitive insulin-secreting beta-like cells
Synthetic biology offers the potential for the design and implementation of rationally designed, complex genetic programmes. Here the authors design a genetic network to trigger the differentiation of patient derived IPSCs into beta-like cells.
- Pratik Saxena
- , Boon Chin Heng
- & Martin Fussenegger
-
Article
| Open Access 3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression
3D replicon distributions arise from stochastic initiation and domino-like DNA replication progression
DNA replication in higher eukaryotes is a complex process occurring in a complex genome environment. Here the authors present a model of DNA replication that incorporates random loop chromatin folding and domino-like fork progression reproducing the spatial and temporal characteristics of S-phase.
- D. Löb
- , N. Lengert
- & B. Drossel
-
Article
| Open Access Engineered kinesin motor proteins amenable to small-molecule inhibition
Engineered kinesin motor proteins amenable to small-molecule inhibition
The use of specific small molecule inhibitors can contribute to the study of kinesins' cellular functions. Here the authors develop a chemical-genetic approach to engineer kinesin motors that can be efficiently inhibited upon addition of cell-permeable molecules.
- Martin F. Engelke
- , Michael Winding
- & Kristen J. Verhey
-
Article
| Open Access Reconstruction and topological characterization of the sigma factor regulatory network of Mycobacterium tuberculosis
Reconstruction and topological characterization of the sigma factor regulatory network of Mycobacterium tuberculosis
Sigma factors are regulatory proteins that reprogram the bacterial RNA polymerase in response to stress conditions to transcribe certain genes, including those for other sigma factors. Here, Chauhan et al. describe the complete sigma factor regulatory network of the pathogen Mycobacterium tuberculosis.
- Rinki Chauhan
- , Janani Ravi
- & Maria Laura Gennaro
-
Article
| Open Access Collaboration between primitive cell membranes and soluble catalysts
Collaboration between primitive cell membranes and soluble catalysts
Early cells likely consisted of fatty acid vesicles enclosing magnesium-dependent ribozymes. Here, the authors show that fatty acid derivatives can form vesicles that, unlike those formed from only unmodified fatty acids, are stable in the presence of magnesium and could support ribozyme catalysis.
- Katarzyna P. Adamala
- , Aaron E. Engelhart
- & Jack W. Szostak
-
Article
| Open Access Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases
Manipulation of prenylation reactions by structure-based engineering of bacterial indolactam prenyltransferases
Regioselective prenylation of small aromatic natural molecules is crucial for their biological activity. Here, the authors present the biochemical and structural characterisation of two prenyltransferases and a structure-based engineering strategy to modulate their substrate specificity.
- Takahiro Mori
- , Lihan Zhang
- & Ikuro Abe
-
Article
| Open Access Rationally engineered Troponin C modulates in vivo cardiac function and performance in health and disease
Rationally engineered Troponin C modulates in vivo cardiac function and performance in health and disease
Heart contraction, which is decreased in disease, is determined by Ca2+binding to troponin C. Here, the authors combine a protein engineering approach with gene therapy to modulate heart contractility in mice with the use of rationally designed Troponin C variants, suggesting a new therapy for diseased hearts.
- Vikram Shettigar
- , Bo Zhang
- & Jonathan P. Davis
-
Article
| Open Access An interactive web-based application for Comprehensive Analysis of RNAi-screen Data
An interactive web-based application for Comprehensive Analysis of RNAi-screen Data
Analysis of RNAi screens is a multi-step process requiring the sequential use of several unrelated resources. Here the authors generate an online resource integrating RNAi analytic tools and filters into a seamless workflow, which improves the specificity, selectivity and reproducibility of the results.
- Bhaskar Dutta
- , Alaleh Azhir
- & Iain D. C. Fraser
-
Article
| Open Access Precision multidimensional assay for high-throughput microRNA drug discovery
Precision multidimensional assay for high-throughput microRNA drug discovery
Progress in drug discovery can be hampered by a limited exploration of chemical space and the difficulty in assessing the full range of drug candidates’ effects on living cells. Here the authors describe a cell-based assay to distinguish between off-target and specific effects of candidate compounds targeting micro RNAs.
- Benjamin Haefliger
- , Laura Prochazka
- & Yaakov Benenson
-
Article
| Open Access Affinity and competition for TBP are molecular determinants of gene expression noise
Affinity and competition for TBP are molecular determinants of gene expression noise
TATA boxes in gene promoters are associated with high level of cell-to-cell variation in gene expression. Through integration of multiple data sets, the authors now provide insights into how the interactions of TBP with DNA and other proteins can lead to noisy expression.
- Charles N. J. Ravarani
- , Guilhem Chalancon
- & M. Madan Babu
-
Article
| Open Access Network-based in silico drug efficacy screening
Network-based in silico drug efficacy screening
Attempts to predict novel use for existing drugs rarely consider information on the impact on the genes perturbed in a given disease. Here, the authors present a novel network-based drug-disease proximity measure that provides insight on gene specific therapeutic effect of drugs and may facilitate drug repurposing.
- Emre Guney
- , Jörg Menche
- & Albert-László Barábasi
-
Article
| Open Access Conversion of graded phosphorylation into switch-like nuclear translocation via autoregulatory mechanisms in ERK signalling
Conversion of graded phosphorylation into switch-like nuclear translocation via autoregulatory mechanisms in ERK signalling
While ERK signalling can produce switch-like cell behaviour, phosphorylation of ERK increases linearly with extracellular signals. Here, the authors solve this seeming contradiction by showing that nuclear translocation of ERK behaves in a switch-like manner and is controlled by ERK activity.
- Yuki Shindo
- , Kazunari Iwamoto
- & Koichi Takahashi
-
Article
| Open Access Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics
Large-scale inference of protein tissue origin in gram-positive sepsis plasma using quantitative targeted proteomics
Sepsis can lead to multiple organ failure that could potentially be reflected by change in plasma protein abundance. Here the authors describe a proteomics strategy that allows the determination of plasma proteins tissue origin in a quantitative manner for use as biomarkers—illustrated in a mouse model of sepsis.
- Erik Malmström
- , Ola Kilsgård
- & Johan Malmström
-
Article
| Open Access Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames
Global proteogenomic analysis of human MHC class I-associated peptides derived from non-canonical reading frames
Cryptic translation of the 'non-coding' genome is increasingly recognised, however its biological significance remains unclear. Laumont et al.employ proteogenomic techniques to map the human immunoproteome, and find that approximately 10% of MHC class I-associated peptides are cryptic.
- Céline M. Laumont
- , Tariq Daouda
- & Claude Perreault
-
Article
| Open Access Proteomic maps of breast cancer subtypes
Proteomic maps of breast cancer subtypes
Breast cancers have been extensively studied at the genomic and transcriptomic levels in the hope of tailoring therapeutic regimens. Here the authors generate deep coverage proteomes from several clinical breast cancer samples, and use machine learning techniques to uncover biological processes altered in specific cancer subtypes.
- Stefka Tyanova
- , Reidar Albrechtsen
- & Tamar Geiger
-
Article
| Open Access Long-term neural and physiological phenotyping of a single human
Long-term neural and physiological phenotyping of a single human
Large-scale, multimodal phenotypic characterisation is a valuable tool to explore brain function. Poldrack et al. collect and relate MRI, psychological, physiological, metabolic and gene expression data from a single human over an 18 month period, providing a rich resource for future studies.
- Russell A. Poldrack
- , Timothy O. Laumann
- & Jeanette A. Mumford
-
Article
| Open Access Single-cell analysis and stochastic modelling unveil large cell-to-cell variability in influenza A virus infection
Single-cell analysis and stochastic modelling unveil large cell-to-cell variability in influenza A virus infection
Cell-to-cell variability in viral infection means that cell population measurements may not be an accurate representation of the process. Using both experimental and modelling approaches the authors confirm this notion showing that influenza virus infections are variable processes affected by intrinsic and extrinsic noise.
- Frank S. Heldt
- , Sascha Y. Kupke
- & Timo Frensing
-
Article
| Open Access Engineering intracellular biomineralization and biosensing by a magnetic protein
Engineering intracellular biomineralization and biosensing by a magnetic protein
Magnetic manipulation of biological systems requires the development of improved molecular handles. Here the authors isolate ferritin mutants with enhanced biomineralization from a yeast genetic screen and show their application to cell separation, multiscale imaging, and construction of sensors.
- Yuri Matsumoto
- , Ritchie Chen
- & Alan Jasanoff
-
Article
| Open Access Topological data analysis for discovery in preclinical spinal cord injury and traumatic brain injury
Topological data analysis for discovery in preclinical spinal cord injury and traumatic brain injury
Data-driven discovery in complex neurological disorders has potential to extract meaningful knowledge from large, heterogeneous datasets. Here the authors apply topological data analysis to assess therapeutic effects in preclinical traumatic brain injury and spinal cord injury research studies.
- Jessica L. Nielson
- , Jesse Paquette
- & Adam R. Ferguson
-
Article
| Open Access Controllability of structural brain networks
Controllability of structural brain networks
Cognitive control is fundamental to human intelligence, yet the principles constraining the neural dynamics of cognitive control remain elusive. Here, the authors use network control theory to demonstrate that the structure of brain networks dictates their functional role in controlling dynamics.
- Shi Gu
- , Fabio Pasqualetti
- & Danielle S. Bassett
-
Article
| Open Access Lower glycolysis carries a higher flux than any biochemically possible alternative
Lower glycolysis carries a higher flux than any biochemically possible alternative
The biochemical pathways of central carbon metabolism are highly conserved across all domains of life. Here, Courtet al. use a computational approach to test all possible pathways of glycolysis and gluconeogenesis and find that the existing trunk pathways may represent a maximal flux solution selected for during evolution.
- Steven J. Court
- , Bartlomiej Waclaw
- & Rosalind J. Allen
-
Article
| Open Access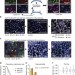 Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method
Highly multiplexed imaging of single cells using a high-throughput cyclic immunofluorescence method
Multiplexed single cell measurements provide insight into connections between cell state and phenotype. Here Lin et al.present CycIF, a high throughput, public domain immunofluorescence method for multiplexed single-cell analysis of adherent cells following live-cell imaging.
- Jia-Ren Lin
- , Mohammad Fallahi-Sichani
- & Peter K. Sorger
-
Article
| Open Access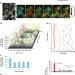 Oscillation of p38 activity controls efficient pro-inflammatory gene expression
Oscillation of p38 activity controls efficient pro-inflammatory gene expression
The prolonged presence of cytokines is necessary to produce a robust pro-inflammatory response through the activation of p38 MAPK. Here, Tomidaet al. show that asynchronous oscillatory activation of p38 MAPK occurs at the single-cell level and is necessary for the proper expression of pro-inflammatory genes.
- Taichiro Tomida
- , Mutsuhiro Takekawa
- & Haruo Saito
-
Article
| Open Access Quantitative analysis reveals how EGFR activation and downregulation are coupled in normal but not in cancer cells
Quantitative analysis reveals how EGFR activation and downregulation are coupled in normal but not in cancer cells
Cells respond to increasing concentrations of EGF by altering the balance between EGFR phosphorylation and ubiquitination. Here the authors show that the establishment of an EGFR signaling threshold requires both a multiplicity of binding sites and cooperative binding of Cbl and Grb2 to the EGFR.
- Fabrizio Capuani
- , Alexia Conte
- & Andrea Ciliberto
-
Article
| Open Access The mechanochemistry of copper reports on the directionality of unfolding in model cupredoxin proteins
The mechanochemistry of copper reports on the directionality of unfolding in model cupredoxin proteins
In metalloproteins, a metal cofactor participates in the formation of the correct fold. Here the authors demonstrate—using single molecule force spectroscopy and the native copper centre as an embedded internal reporter—that the blue-copper proteins azurin and plastocyanin unfold via two independent competing pathways under force.
- Amy E. M. Beedle
- , Ainhoa Lezamiz
- & Sergi Garcia-Manyes
-
Article
| Open Access Phosphoproteomics reveals malaria parasite Protein Kinase G as a signalling hub regulating egress and invasion
Phosphoproteomics reveals malaria parasite Protein Kinase G as a signalling hub regulating egress and invasion
Protein kinases expressed by the malaria parasite Plasmodium falciparum represent potentially valuable drug targets. Alam et al. identify proteins whose phosphorylation is dependent on the kinase PfPKG, revealing a regulatory network controlling parasite egress from red blood cells and calcium signalling.
- Mahmood M. Alam
- , Lev Solyakov
- & Andrew B. Tobin
-
Article |
 Allostery through the computational microscope: cAMP activation of a canonical signalling domain
Allostery through the computational microscope: cAMP activation of a canonical signalling domain
Allostery, communication between distant parts of a protein, is a key element of enzyme catalysis. Here the authors combine existing experimental data with molecular dynamics simulations and Markov state models to provide insight into the mechanism of ligand-induced allostery within the cyclicnucleotide-binding domain of protein kinase A.
- Robert D. Malmstrom
- , Alexandr P. Kornev
- & Rommie E. Amaro
-
Article
| Open Access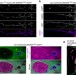 Cell-selective labelling of proteomes in Drosophila melanogaster
Cell-selective labelling of proteomes in Drosophila melanogaster
Mutated tRNA synthetases can incorporate non-canonical amino acids into proteins. Erdmann et al. exploit this property to metabolically label newly synthesized proteins in selected cell types in Drosophila, and demonstrate their detection using proteomics (BONCAT) and fluorescence imaging (FUNCAT).
- Ines Erdmann
- , Kathrin Marter
- & Daniela C. Dieterich
-
Article
| Open Access Impaired protein translation in Drosophila models for Charcot–Marie–Tooth neuropathy caused by mutant tRNA synthetases
Impaired protein translation in Drosophila models for Charcot–Marie–Tooth neuropathy caused by mutant tRNA synthetases
Charcot–Marie–Tooth (CMT) neuropathy is associated with dominant mutations in five tRNA synthetase genes. Niehues et al. use BONCAT and FUNCAT to monitor proteome dynamics in a DrosophilaCMT model, and reveal that these mutations result in translational slowdown.
- Sven Niehues
- , Julia Bussmann
- & Erik Storkebaum
-
Article
| Open Access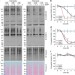 System-wide identification of wild-type SUMO-2 conjugation sites
System-wide identification of wild-type SUMO-2 conjugation sites
Tryptic digestion of SUMOylated proteins generates large peptides, rendering proteomic characterisation of this post-translational modification particularly challenging unless mutant SUMO is used. Hendriks et al.present a method that allows the quantitative identification of wild-type SUMO sites.
- Ivo A. Hendriks
- , Rochelle C. D’Souza
- & Alfred C. O. Vertegaal
-
Article
| Open Access Extreme multifunctional proteins identified from a human protein interaction network
Extreme multifunctional proteins identified from a human protein interaction network
Proteins are sometimes implicated in separate and seemingly unrelated processes, so called moonlighting functions. Here the authors use bioinformatics tools to identify extreme multifunctional proteins and define a signature of extreme multifunctionality.
- Charles E. Chapple
- , Benoit Robisson
- & Christine Brun
-
Article
| Open Access Combinatorial code governing cellular responses to complex stimuli
Combinatorial code governing cellular responses to complex stimuli
Cells constantly integrate information from multiple stimuli. By considering every possible means by which two stimuli can interact, Cappuccio et al. define 10 interaction modes and demonstrate their preferential use by dendritic cells responding to different combinations of microbial and host inflammatory cues.
- Antonio Cappuccio
- , Raphaël Zollinger
- & Vassili Soumelis
-
Article
| Open Access Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics
Large-scale determination of absolute phosphorylation stoichiometries in human cells by motif-targeting quantitative proteomics
Measuring phosphorylation stoichiometry on a proteomic scale remains a challenge. Tsai et al. develop a technique to measure the basal level of phosphorylation stoichiometry in a single human phosphoproteome and identify molecular changes associated with gefitinib resistance in lung cancer cells.
- Chia-Feng Tsai
- , Yi-Ting Wang
- & Yu-Ju Chen
Browse broader subjects
Browse narrower subjects
- Bayesian inference
- Biobricks
- Biochemical networks
- Bioenergetics
- Cellular noise
- Complexity
- Computer modelling
- Computer science
- Control theory
- Criticality
- Differential equations
- DNA computing and cryptography
- Dynamic networks
- Dynamical systems
- Emergence
- Evolvability
- Genetic circuit engineering
- Genetic interaction
- Genomic engineering
- Information theory
- Logic gates
- Metabolic engineering
- Modularity
- Molecular engineering
- Molecular fluctuations
- Multicellular systems
- Multistability
- Nonlinear dynamics
- Numerical simulations
- Oscillators
- Population dynamics
- Programming language
- Protein engineering
- Regulatory networks
- Reverse engineering
- Robustness
- Signal processing
- Single-cell imaging
- Software
- Standardization
- Stochastic modelling
- Stochastic networks
- Synthetic biology
- Systems analysis
- Time series

 Glycolytic regulation of cell rearrangement in angiogenesis
Glycolytic regulation of cell rearrangement in angiogenesis