Featured
-
-
Letter |
 Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans
Terminal Pleistocene Alaskan genome reveals first founding population of Native Americans
An Ancient Beringian population from Late Pleistocene Alaska and the ancestors of other Native American groups descended from a single founding population that diversified around twenty thousand years ago.
- J. Víctor Moreno-Mayar
- , Ben A. Potter
- & Eske Willerslev
-
Letter |
 Genetic diversity of the African malaria vector Anopheles gambiae
Genetic diversity of the African malaria vector Anopheles gambiae
Genome sequencing analyses from 765 specimens of Anopheles gambiae and Anopheles coluzzii from 15 locations across Africa characterize patterns of gene flow and variations in population size, and provide a resource for studying the evolution of natural malaria vector populations.
- Alistair Miles
- , Nicholas J. Harding
- & Dominic P. Kwiatkowski
-
Letter |
 Parallel palaeogenomic transects reveal complex genetic history of early European farmers
Parallel palaeogenomic transects reveal complex genetic history of early European farmers
In European Neolithic populations, the arrival of farmers prompted admixture with local hunter-gatherers over many centuries, resulting in distinct signatures in each region due to a complex series of interactions.
- Mark Lipson
- , Anna Szécsényi-Nagy
- & David Reich
-
Letter |
 Detecting evolutionary forces in language change
Detecting evolutionary forces in language change
Analyses of digital corpora of annotated texts reveal the influence of stochastic drift versus selection in grammatical shifts in English and provide a general method for quantitatively testing theories of language change.
- Mitchell G. Newberry
- , Christopher A. Ahern
- & Joshua B. Plotkin
-
Letter |
 Parental influence on human germline de novo mutations in 1,548 trios from Iceland
Parental influence on human germline de novo mutations in 1,548 trios from Iceland
Whole-genome sequencing data of 14,688 Icelanders, including 1,548 parent–offspring trios, show how the age and sex of parents affect the rate and spectrum of de novo mutations.
- Hákon Jónsson
- , Patrick Sulem
- & Kari Stefansson
-
Letter |
 Genetic origins of the Minoans and Mycenaeans
Genetic origins of the Minoans and Mycenaeans
New genome-wide data for ancient, Bronze Age individuals, including Minoans, Mycenaeans, and southwestern Anatolians, show that Minoans and Mycenaeans were genetically very similar yet distinct, supporting the idea of continuity but not isolation in the history of populations of the Aegean.
- Iosif Lazaridis
- , Alissa Mittnik
- & George Stamatoyannopoulos
-
Letter
| Open Access Sequencing and de novo assembly of 150 genomes from Denmark as a population reference
Sequencing and de novo assembly of 150 genomes from Denmark as a population reference
A report of high-depth, short-read sequencing and de novo assemblies for 150 individuals from 50 parent–offspring trios as part of establishing a population reference genome for the GenomeDenmark project.
- Lasse Maretty
- , Jacob Malte Jensen
- & Mikkel Heide Schierup
-
Article |
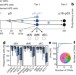 Common genetic variation drives molecular heterogeneity in human iPSCs
Common genetic variation drives molecular heterogeneity in human iPSCs
Genetic and phenotypic analysis reveals expression quantitative trait loci in human induced pluripotent stem cell lines associated with cancer and disease.
- Helena Kilpinen
- , Angela Goncalves
- & Daniel J. Gaffney
-
Letter |
 Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity
Human knockouts and phenotypic analysis in a cohort with a high rate of consanguinity
By sequencing the exomes of 10,503 individuals living in Pakistan, the authors identify rare predicted loss-of-function mutations that are estimated to knock out genes and correlate these mutations with a broad range of phenotypes, providing a framework for a human knockout project.
- Danish Saleheen
- , Pradeep Natarajan
- & Sekar Kathiresan
-
Article |
 Aboriginal mitogenomes reveal 50,000 years of regionalism in Australia
Aboriginal mitogenomes reveal 50,000 years of regionalism in Australia
Analysis of Aboriginal Australian mitochondrial genomes shows geographic patterns and deep splits across the major haplogroups that indicate a single, rapid migration along the coasts around 49–45 ka, followed by longstanding persistence in discrete geographic areas.
- Ray Tobler
- , Adam Rohrlach
- & Alan Cooper
-
Article |
 Balancing selection shapes density-dependent foraging behaviour
Balancing selection shapes density-dependent foraging behaviour
Natural isolates of Caenorhabditis elegans nematodes differ in their sensitivity to the anti-exploratory pheromone icas#9, yielding two distinct foraging strategies that possess different survival advantages depending on environmental conditions such as food distribution.
- Joshua S. Greene
- , Maximillian Brown
- & Cornelia I. Bargmann
-
Letter |
 Genomic insights into the peopling of the Southwest Pacific
Genomic insights into the peopling of the Southwest Pacific
Analysis of ancient DNA from four individuals who lived in Vanuatu and Tonga between 2,300 and 3,100 years ago suggests that the Papuan ancestry seen in present-day occupants of this region was introduced at a later date.
- Pontus Skoglund
- , Cosimo Posth
- & David Reich
-
Article |
 A genomic history of Aboriginal Australia
A genomic history of Aboriginal Australia
Whole-genome sequence data for 108 individuals representing 28 language groups across Australia and five language groups for Papua New Guinea suggests that Aboriginal Australians and Papuans diverged from Eurasian populations approximately 60–100 thousand years ago, following a single out-of-Africa dispersal and subsequent admixture with archaic populations.
- Anna-Sapfo Malaspinas
- , Michael C. Westaway
- & Eske Willerslev
-
Article |
 Genomic insights into the origin of farming in the ancient Near East
Genomic insights into the origin of farming in the ancient Near East
Analysis of DNA from ancient individuals of the Near East documents the extreme substructure among the populations which transitioned to farming, a structure that was maintained throughout the transition from hunter–gatherer to farmer but that broke down over the next five thousand years.
- Iosif Lazaridis
- , Dani Nadel
- & David Reich
-
Article |
 The genetic history of Ice Age Europe
The genetic history of Ice Age Europe
Analysis of ancient genomic data of 51 humans from Eurasia dating from 45,000 to 7,000 years ago provides insight into the population history of pre-Neolithic Europe and support for recurring migration and population turnover in Europe during this period.
- Qiaomei Fu
- , Cosimo Posth
- & David Reich
-
Article |
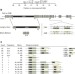 Schizophrenia risk from complex variation of complement component 4
Schizophrenia risk from complex variation of complement component 4
WebSchizophrenia is associated with genetic variation at the major histocompatibility complex locus; this study reveals that alleles at this locus associate with schizophrenia in proportion to their tendency to generate greater expression of complement component 4 (C4A) genes and that C4 promotes the elimination of synpases.
- Aswin Sekar
- , Allison R. Bialas
- & Steven A. McCarroll
-
Article |
 Genome-wide patterns of selection in 230 ancient Eurasians
Genome-wide patterns of selection in 230 ancient Eurasians
The first genome-wide scan for selection using ancient DNA, based on data from 230 West Eurasians dating between to 6500 and 300 bc and including new data from 163 individuals among which are 26 Neolithic Anatolians, provides a direct view of selection on loci associated with diet, pigmentation and immunity.
- Iain Mathieson
- , Iosif Lazaridis
- & David Reich
-
Article
| Open Access A global reference for human genetic variation
A global reference for human genetic variation
Results for the final phase of the 1000 Genomes Project are presented including whole-genome sequencing, targeted exome sequencing, and genotyping on high-density SNP arrays for 2,504 individuals across 26 populations, providing a global reference data set to support biomedical genetics.
- Adam Auton
- , Gonçalo R. Abecasis
- & Gonçalo R. Abecasis
-
Letter |
 A novel locus of resistance to severe malaria in a region of ancient balancing selection
A novel locus of resistance to severe malaria in a region of ancient balancing selection
A multi-centre genome-wide association study of severe malaria in African children uncovers a new resistance locus close to a cluster of genes encoding glycophorins, which are receptors used by the malaria-causing parasite to invade red blood cells.
- Gavin Band
- , Kirk A. Rockett
- & Victoria Cornelius
-
Letter |
 Genetic evidence for two founding populations of the Americas
Genetic evidence for two founding populations of the Americas
Previous genetic studies have suggested that the Americas were peopled by a single founding population of Eurasian origin, but a genome-wide study of 30 Native American groups shows that Amazonian Native Americans also have a second source of ancestry that is deeply related to indigenous Australians, New Guineans and Andaman Islanders.
- Pontus Skoglund
- , Swapan Mallick
- & David Reich
-
Letter
| Open Access The ancestry and affiliations of Kennewick Man
The ancestry and affiliations of Kennewick Man
Kennewick Man, a 8,500-year-old male human skeleton discovered in Washington state, USA, has been the subject of scientific and legal controversy; here a DNA analysis shows that Kennewick Man is closer to modern Native Americans than to any other extant population worldwide.
- Morten Rasmussen
- , Martin Sikora
- & Eske Willerslev
-
Article |
 Population genomics of Bronze Age Eurasia
Population genomics of Bronze Age Eurasia
An analysis of 101 ancient human genomes from the Bronze Age (3000–1000 bc) reveals large-scale population migrations in Eurasia consistent with the spread of Indo-European languages; individuals frequently had light skin pigmentation but were not lactose tolerant.
- Morten E. Allentoft
- , Martin Sikora
- & Eske Willerslev
-
Article |
 The fine-scale genetic structure of the British population
The fine-scale genetic structure of the British population
Extensive genetic analysis of over 2,000 individuals from different locations in Britain reveals striking fine-scale patterns of population structure; comparisons with similar genetic data from the European continent reveal the legacy of earlier population migrations and information about the ancestry of current populations in specific geographic regions.
- Stephen Leslie
- , Bruce Winney
- & Walter Bodmer
-
Letter |
 Selection on noise constrains variation in a eukaryotic promoter
Selection on noise constrains variation in a eukaryotic promoter
Quantifying activity of cis-regulatory sequences controlling gene expression shows that selection on expression noise has a greater impact on sequence variation than selection on mean expression level.
- Brian P. H. Metzger
- , David C. Yuan
- & Patricia J. Wittkopp
-
Letter |
 Massive migration from the steppe was a source for Indo-European languages in Europe
Massive migration from the steppe was a source for Indo-European languages in Europe
A genome-wide analysis of 69 ancient Europeans reveals the history of population migrations around the time that Indo-European languages arose in Europe, when there was a large migration into Europe from the Eurasian steppe in the east (providing a genetic ancestry still present in Europeans today); these findings support a ‘steppe origin’ hypothesis for how some Indo-European languages arose.
- Wolfgang Haak
- , Iosif Lazaridis
- & David Reich
-
Article
| Open Access The African Genome Variation Project shapes medical genetics in Africa
The African Genome Variation Project shapes medical genetics in Africa
The African Genome Variation Project contains the whole-genome sequences of 320 individuals and dense genotypes on 1,481 individuals from sub-Saharan Africa; it enables the design and interpretation of genomic studies, with implications for finding disease loci and clues to human origins.
- Deepti Gurdasani
- , Tommy Carstensen
- & Manjinder S. Sandhu
-
Article |
 The genetics of monarch butterfly migration and warning colouration
The genetics of monarch butterfly migration and warning colouration
The monarch butterfly, well known for its spectacular annual migration across North America, is shown by genome sequencing of monarchs from around the world to have been ancestrally migratory and to have dispersed out of North America to occupy its current broad distribution; the authors also discovered signatures of selection associated with migration within loci implicated in flight muscle function, leading to greater flight efficiency.
- Shuai Zhan
- , Wei Zhang
- & Marcus R. Kronforst
-
Letter |
 Comparative population genomics in animals uncovers the determinants of genetic diversity
Comparative population genomics in animals uncovers the determinants of genetic diversity
Genome-wide DNA polymorphism analysis across 76 animal species reveals a strong effect of ecological strategies, and particularly parental investment, on species levels of genetic diversity.
- J. Romiguier
- , P. Gayral
- & N. Galtier
-
Letter |
 Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA
Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA
Admixture with other hominin species helped humans to adapt to high-altitude environments; the EPAS1 gene in Tibetan individuals has an unusual haplotype structure that probably resulted from introgression of DNA from Denisovan or Denisovan-related individuals into humans, and this haplotype is only found in Denisovans and Tibetans, and at low frequency among Han Chinese.
- Emilia Huerta-Sánchez
- , Xin Jin
- & Rasmus Nielsen
-
Letter |
 doublesex is a mimicry supergene
doublesex is a mimicry supergene
The phenomenon of sex-limited mimicry is phylogenetically widespread in the swallowtail butterfly genus Papilio — now, a single gene, doublesex, is shown to control supergene mimicry, a finding that is in contrast to the long-held view that supergenes are likely to be controlled by a tightly linked cluster of loci.
- K. Kunte
- , W. Zhang
- & M. R. Kronforst
-
Letter |
 Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico
Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico
A risk haplotype for type 2 diabetes is identified with four amino acid substitutions in SLC16A11, which is present at ∼50% frequency in Native American samples and ∼10% in east Asian samples, but is rare in European and African samples; SLC16A11 may alter hepatic lipid metabolism, causing an increase in triacylglycerol levels.
- Amy L. Williams
- , Suzanne B. R. Jacobs
- & Teresa Tusié-Luna
-
Letter |
 Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans
Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans
Draft genomes of two south-central Siberian individuals dating to 24,000 and 17,000 years ago show that they are genetically closely related to modern-day western Eurasians and Native Americans but not to east Asians; the results have implications for our understanding of the origins of Native Americans.
- Maanasa Raghavan
- , Pontus Skoglund
- & Eske Willerslev
-
Article |
 Transcriptome and genome sequencing uncovers functional variation in humans
Transcriptome and genome sequencing uncovers functional variation in humans
Sequencing and deep analysis of mRNA and miRNA from lymphoblastoid cell lines of 462 individuals from the 1000 Genomes Project reveal widespread genetic variation affecting the regulation of most genes, with transcript structure and expression level variation being equally common but genetically largely independent, and the analyses point to putative causal variants for dozens of disease-associated loci.
- Tuuli Lappalainen
- , Michael Sammeth
- & Emmanouil T. Dermitzakis
-
Brief Communications Arising |
 Breen et al. reply
Breen et al. reply
- Michael S. Breen
- , Carsten Kemena
- & Fyodor A. Kondrashov
-
Brief Communications Arising |
 The role of epistasis in protein evolution
The role of epistasis in protein evolution
- David M. McCandlish
- , Etienne Rajon
- & Joshua B. Plotkin
-
Letter |
 The genomic signature of dog domestication reveals adaptation to a starch-rich diet
The genomic signature of dog domestication reveals adaptation to a starch-rich diet
Whole-genome resequencing of dogs and wolves helps identify genomic regions that are likely to represent targets for selection during dog domestication.
- Erik Axelsson
- , Abhirami Ratnakumar
- & Kerstin Lindblad-Toh
-
Research Highlights |
 Romani have Indian ancestry
Romani have Indian ancestry
-
Research Highlights |
 Nomadic group of Irish descent
Nomadic group of Irish descent
-
Article
| Open Access An integrated map of genetic variation from 1,092 human genomes
An integrated map of genetic variation from 1,092 human genomes
This report from the 1000 Genomes Project describes the genomes of 1,092 individuals from 14 human populations, providing a resource for common and low-frequency variant analysis in individuals from diverse populations; hundreds of rare non-coding variants at conserved sites, such as motif-disrupting changes in transcription-factor-binding sites, can be found in each individual.
- Gil A. McVean
- , David M. Altshuler (Co-Chair)
- & Gil A. McVean
-
Editorial |
Fighting chance
Collaboration between geneticists and economists has the potential to bear fruit.
-
News |
 Economics and genetics meet in uneasy union
Economics and genetics meet in uneasy union
Use of population-genetic data to predict economic success sparks war of words.
- Ewen Callaway
-
Article
| Open Access A map of rice genome variation reveals the origin of cultivated rice
A map of rice genome variation reveals the origin of cultivated rice
Whole-genome sequences of wild rice and cultivated rice varieties are used to produce a map of rice genome variation, and show that rice was probably first domesticated in southern China.
- Xuehui Huang
- , Nori Kurata
- & Bin Han
-
News |
 African neighbours divided by their genes
African neighbours divided by their genes
Geographically close human populations in southern Africa have been genetically isolated for thousands of years.
- Matt Kaplan
-
Letter |
 FTO genotype is associated with phenotypic variability of body mass index
FTO genotype is associated with phenotypic variability of body mass index
A meta-analysis of genome-wide association studies of phenotypic variation for height and body mass index in human populations using 170,000 samples shows that one single nucleotide polymorphism at the FTO locus, which is associated with obesity, is also associated with phenotypic variation.
- Jian Yang
- , Ruth J. F. Loos
- & Peter M. Visscher
-
News |
 Soapy taste of coriander linked to genetic variants
Soapy taste of coriander linked to genetic variants
Dislike of herb traced to genes encoding odour and taste receptors.
- Ewen Callaway
-
Article
| Open Access The accessible chromatin landscape of the human genome
The accessible chromatin landscape of the human genome
An extensive map of human DNase I hypersensitive sites, markers of regulatory DNA, in 125 diverse cell and tissue types is described; integration of this information with other ENCODE-generated data sets identifies new relationships between chromatin accessibility, transcription, DNA methylation and regulatory factor occupancy patterns.
- Robert E. Thurman
- , Eric Rynes
- & John A. Stamatoyannopoulos
-
News Feature |
 Science on the Silk Road: Taste for adventure
Science on the Silk Road: Taste for adventure
Scientists have travelled the ancient Silk Road to understand how genes shape people's love for foods.
- Alison Abbott
-
Research Highlights |
 Hunter-gatherer genes
Hunter-gatherer genes
-
News |
 Hunter-gatherer genomes a trove of genetic diversity
Hunter-gatherer genomes a trove of genetic diversity
Sequences from African groups offer genetic clues about disease, height and ancient human breeding.
- Ewen Callaway

 The Beaker phenomenon and the genomic transformation of northwest Europe
The Beaker phenomenon and the genomic transformation of northwest Europe