Featured
-
-
Letter |
 Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration
Pif1 helicase and Polδ promote recombination-coupled DNA synthesis via bubble migration
This paper demonstrates that Pif1 helicase works with polymerase d to promote DNA synthesis through a migrating D-loop, a mechanism used to copy tens of kilobases during repair of chromosome breaks by break-induced replication (BIR).
- Marenda A. Wilson
- , YoungHo Kwon
- & Grzegorz Ira
-
Letter |
 Migrating bubble during break-induced replication drives conservative DNA synthesis
Migrating bubble during break-induced replication drives conservative DNA synthesis
This paper demonstrates that the mechanism of break-induced replication (BIR) is significantly different from S-phase replication, as it proceeds via a migrating bubble driven by Pif1 helicase, results in conservative inheritance of newly synthesized DNA, and is inherently mutagenic.
- Natalie Saini
- , Sreejith Ramakrishnan
- & Anna Malkova
-
Letter |
 Avoiding chromosome pathology when replication forks collide
Avoiding chromosome pathology when replication forks collide
The site of collision between two chromosome replication forks can be used to reinitiate replication independent of an active origin, with potentially pathogenic effects.
- Christian J. Rudolph
- , Amy L. Upton
- & Robert G. Lloyd
-
Article |
 ATPase-dependent quality control of DNA replication origin licensing
ATPase-dependent quality control of DNA replication origin licensing
The authors describe how the eukaryotic replicative helicase is recruited to origins and reveal a novel ATPase-dependent quality control mechanism.
- Jordi Frigola
- , Dirk Remus
- & John F. X. Diffley
-
Letter |
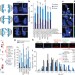 Replication stress links structural and numerical cancer chromosomal instability
Replication stress links structural and numerical cancer chromosomal instability
A mechanism to explain chromosomal instability (CIN) in colorectal cancer is demonstrated; three new CIN-suppressor genes (PIGN, MEX3C and ZNF516) encoded on chromosome 18q are identified, the loss of which leads to DNA replication stress, resulting in structural and numerical chromosome segregation errors, which are shown to be identical to phenotypes seen in CIN cells.
- Rebecca A. Burrell
- , Sarah E. McClelland
- & Charles Swanton
-
News & Views |
 Molecular hurdles cleared with ease
Molecular hurdles cleared with ease
Single-molecule studies reveal that a ring-like enzyme that encircles and 'slides' along one strand of duplex DNA, separating it from the other strand, overcomes molecular barriers in its path by transiently opening its ring. See Article p.205
- Michael A. Trakselis
- & Brian W. Graham
-
Article |
 Bypass of a protein barrier by a replicative DNA helicase
Bypass of a protein barrier by a replicative DNA helicase
Single-molecule and ensemble assays are used to show that large T antigen, the replicative DNA helicase of the simian virus 40 (SV40), unwinds DNA as a single hexamer by steric exclusion and is able to bypass covalent DNA–protein crosslinks.
- Hasan Yardimci
- , Xindan Wang
- & Johannes C. Walter
-
Letter |
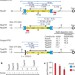 Recombination-restarted replication makes inverted chromosome fusions at inverted repeats
Recombination-restarted replication makes inverted chromosome fusions at inverted repeats
A new mechanism of chromosomal rearrangement is identified through the observation that broken or collapsed DNA replication forks restarted by homologous recombination have a high propensity for U-turns at short inverted repeats; the error-prone nature of this mechanism is suggested to contribute to gross chromosomal rearrangements and copy-number variations present in cancer and other genomic disorders.
- Ken’Ichi Mizuno
- , Izumi Miyabe
- & Johanne M. Murray
-
Letter |
 Coordinated control of replication and transcription by a SAPK protects genomic integrity
Coordinated control of replication and transcription by a SAPK protects genomic integrity
Upregulation of gene transcription in stressed cells can lead to clashes between the transcription and repair machineries; here, a stress-activated protein kinase (SAPK), Hog1, is shown to coordinate these two processes in yeast.
- Alba Duch
- , Irene Felipe-Abrio
- & Francesc Posas
-
Letter |
 DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9
DNA-repair scaffolds dampen checkpoint signalling by counteracting the adaptor Rad9
DNA damage or replication stress induces the activation of checkpoint kinases, pausing the cell cycle so that DNA repair can take place; checkpoint activation must be regulated to prevent the cell-cycle arrest from persisting after damage is repaired, and now the Slx4–Rtt107 complex is shown to regulate checkpoint kinase activity by directly monitoring DNA-damage signalling.
- Patrice Y. Ohouo
- , Francisco M. Bastos de Oliveira
- & Marcus B. Smolka
-
News & Views |
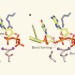 DNA replication caught in the act
DNA replication caught in the act
By freezing a DNA polymerase enzyme at several points along its reaction pathway, a sequence of X-ray crystal structures has been obtained, showing how the enzyme replicates DNA and revealing surprising mechanistic details. See Article p.196
- Kenneth A. Johnson
-
Letter |
 The human CST complex is a terminator of telomerase activity
The human CST complex is a terminator of telomerase activity
The human CST complex is shown to interact with the telomeric primer and the POT1–TPP1 complex to inhibit telomerase activity in late S phase, thereby keeping unrestrained telomere lengthening in check.
- Liuh-Yow Chen
- , Sophie Redon
- & Joachim Lingner
-
News & Views |
 How to duplicate a DNA package
How to duplicate a DNA package
Cells replicate half of their genome as short fragments that are put together later on. The way in which this process is linked to the formation of DNA–protein complexes called nucleosomes is now becoming clearer. See Article p.434
- Alysia Vandenberg
- & Geneviève Almouzni
-
Article |
 Intrinsic coupling of lagging-strand synthesis to chromatin assembly
Intrinsic coupling of lagging-strand synthesis to chromatin assembly
Genome-wide deep sequencing of Okazaki fragments in S. cerevisae reveals a connection between lagging-strand synthesis and chromatin assembly.
- Duncan J. Smith
- & Iestyn Whitehouse
-
Letter |
 The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier–Gorlin syndrome
The BAH domain of ORC1 links H4K20me2 to DNA replication licensing and Meier–Gorlin syndrome
The ORC1 BAH domain is shown to be a module that recognizes a histone modification associated with replication origins, providing insight into the aetiology of Meier–Gorlin syndrome.
- Alex J. Kuo
- , Jikui Song
- & Or Gozani
-
Letter |
 RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II
RNAi promotes heterochromatic silencing through replication-coupled release of RNA Pol II
- Mikel Zaratiegui
- , Stephane E. Castel
- & Robert A. Martienssen
-
Letter |
 Self-replication of information-bearing nanoscale patterns
Self-replication of information-bearing nanoscale patterns
- Tong Wang
- , Ruojie Sha
- & Nadrian C. Seeman
-
Article |
 DNA stretching by bacterial initiators promotes replication origin opening
DNA stretching by bacterial initiators promotes replication origin opening
- Karl E. Duderstadt
- , Kevin Chuang
- & James M. Berger
-
Research Highlights |
 DNA replication catastrophe
DNA replication catastrophe
-
Letter |
 ATP-induced helicase slippage reveals highly coordinated subunits
ATP-induced helicase slippage reveals highly coordinated subunits
- Bo Sun
- , Daniel S. Johnson
- & Michelle D. Wang
-
Letter |
 Coordination of DNA replication and histone modification by the Rik1–Dos2 complex
Coordination of DNA replication and histone modification by the Rik1–Dos2 complex
- Fei Li
- , Rob Martienssen
- & W. Zacheus Cande
-
Letter |
 Chromosome length influences replication-induced topological stress
Chromosome length influences replication-induced topological stress
During replication, topological stress builds ahead of the polymerase. Current models propose that linear eukaryotic chromosomes are divided into topological domains, and that stress is relieved by the activity of a topoisomerase. Here, it is found that replication stress seems to be present throughout the chromosome, rather than in domains, and that the relief of stress in longer chromosomes is facilitated by the activity of the cohesin/condensin-like Smc5/6 complex as well as by topoisomerase. They propose that the Smc5/6 complex prevent formation of topological tension ahead of the replication fork by promoting fork rotation, leading to the formation of sister chromatin intertwinings behind.
- Andreas Kegel
- , Hanna Betts-Lindroos
- & Camilla Sjögren
-
Letter |
 Co-directional replication–transcription conflicts lead to replication restart
Co-directional replication–transcription conflicts lead to replication restart
As the rates of replication and transcription are different, the machineries that carry out these processes are bound to clash on DNA. In contrast to results from head-on collisions, co-directional encounters have been shown to have mild effects in vitro, requiring no additional replication restart factors. It is now shown that in bacterial cells, both types of events require the activities of restart proteins to resume replication when a transcription complex is encountered.
- Houra Merrikh
- , Cristina Machón
- & Panos Soultanas
-
News & Views |
 DNA fragility put into context
DNA fragility put into context
Fragile sites are genomic regions prone to deletions or other alterations during DNA replication. The reason for the susceptibility of these sites to damage may be simple: they contain few replication initiation sites. See Letter p.120
- Kay Huebner
-
Letter |
 CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR
CENP-B preserves genome integrity at replication forks paused by retrotransposon LTR
Studies have indicated an undefined role in DNA replication for CENP-B, a DNA binding protein associated with heterochromatin, centromeres and retrotransposon long terminal repeats (LTRs). Here it is shown that Sap1, which binds LTRs, promotes genomic instability when CENP-B activity is absent. CENP-B facilitates replication fork progression through LTRs in a way that protects against rearrangements.
- Mikel Zaratiegui
- , Matthew W. Vaughn
- & Robert A. Martienssen
-
Research Highlights |
Biophysics: DNA replication control
-
Letter |
 Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation
Checkpoint-dependent inhibition of DNA replication initiation by Sld3 and Dbf4 phosphorylation
Two classes of enzyme — cyclin-dependent kinases (CDK) and Dbf4-dependent kinase (DDK) — facilitate the initiation of DNA replication in eukaryotes. It is now shown that, when DNA damage is sensed, another kinase, Rad53, halts the firing of late replication origins by inhibiting both the CDK and the DDK pathways. Rad53 acts on DDK directly by inhibiting Dbf4, whereas the CDK pathway is blocked by Rad53-mediated phosphorylation of the downstream CDK substrate Sld3.
- Philip Zegerman
- & John F. X. Diffley
-
Letter |
 Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases
Regulation of heterochromatic DNA replication by histone H3 lysine 27 methyltransferases
DNA replication occurs only once per cell cycle, and numerous pathways prevent re-replication. Here it is shown that mutations in ARABIDOPSIS TRITHORAX-RELATED PROTEIN5 (ATXR5) and ATXR6 — which encode histone methyltransferases — lead to re-replication of specific genomic locations, notably those corresponding to transposons and other repetitive and silenced elements. ATXR5 and ATXR6 are proposed to be components of a pathway that prevents over-replication of heterochromatin in Arabidopsis.
- Yannick Jacob
- , Hume Stroud
- & Steven E. Jacobsen
-
Letter |
 Ubiquitin-dependent DNA damage bypass is separable from genome replication
Ubiquitin-dependent DNA damage bypass is separable from genome replication
Post-replicative repair (PRR) enables cells to bypass or overcome DNA damage during DNA replication. In eukaryotes, ubiquitylation of the replication clamp PCNA by components of the RAD6 pathway activates damage bypass. When this occurs has been debated. It is now shown that PRR can be postponed until much of the undamaged genome is replicated. Moreover, it seems that PRR occurs mainly by an error-prone process, with error-free bypass playing a minor role.
- Yasukazu Daigaku
- , Adelina A. Davies
- & Helle D. Ulrich
-
Letter |
 The Dbf4–Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4
The Dbf4–Cdc7 kinase promotes S phase by alleviating an inhibitory activity in Mcm4
Kinase regulatory pathways are used in eukaryotic DNA replication to facilitate coordination with other processes during cell division cycles and response to environmental cues. The Dbf4–Cdc7 kinase (DDK) is one of at least two cell-cycle-regulated protein kinase systems essential for initiation of DNA replication. DDK is now shown to relieve the inhibitory activity of the amino-terminal domain of the replicative helicase Mcm4, thus promoting S phase.
- Yi-Jun Sheu
- & Bruce Stillman

 Accelerated growth in the absence of DNA replication origins
Accelerated growth in the absence of DNA replication origins