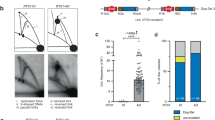
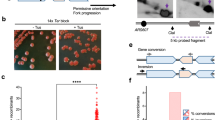
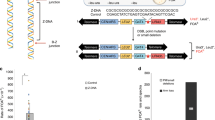
Abstract
Impediments to DNA replication are known to induce gross chromosomal rearrangements (GCRs) and copy-number variations (CNVs). GCRs and CNVs underlie human genomic disorders1 and are a feature of cancer2. During cancer development, environmental factors and oncogene-driven proliferation promote replication stress. Resulting GCRs and CNVs are proposed to contribute to cancer development and therapy resistance3. When stress arrests replication, the replisome remains associated with the fork DNA (stalled fork) and is protected by the inter-S-phase checkpoint. Stalled forks efficiently resume when the stress is relieved. However, if the polymerases dissociate from the fork (fork collapse) or the fork structure breaks (broken fork), replication restart can proceed either by homologous recombination or microhomology-primed re-initiation4,5. Here we ascertain the consequences of replication with a fork restarted by homologous recombination in fission yeast. We identify a new mechanism of chromosomal rearrangement through the observation that recombination-restarted forks have a considerably high propensity to execute a U-turn at small inverted repeats (up to 1 in 40 replication events). We propose that the error-prone nature of restarted forks contributes to the generation of GCRs and gene amplification in cancer, and to non-recurrent CNVs in genomic disorders.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lupski, J. R. Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet. 14, 417–422 (1998)
Liu, P., Carvalho, C. M., Hastings, P. & Lupski, J. R. Mechanisms for recurrent and complex human genomic rearrangements. Curr. Opin. Genet. Dev. 22, 211–220 (2012)
Mondello, C., Smirnova, A. & Giulotto, E. Gene amplification, radiation sensitivity and DNA double-strand breaks. Mutat. Res. 704, 29–37 (2010)
Lee, J. A., Carvalho, C. M. & Lupski, J. R. A. DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131, 1235–1247 (2007)
Hastings, P. J., Ira, G. & Lupski, J. R. A microhomology-mediated break-induced replication model for the origin of human copy number variation. PLoS Genet. 5, e1000327 (2009)
Petermann, E. & Helleday, T. Pathways of mammalian replication fork restart. Nature Rev. Mol. Cell Biol. 11, 683–687 (2010)
Blow, J. J., Ge, X. Q. & Jackson, D. A. How dormant origins promote complete genome replication. Trends Biochem. Sci. 36, 405–414 (2011)
Letessier, A. et al. Cell-type-specific replication initiation programs set fragility of the FRA3B fragile site. Nature 470, 120–123 (2011)
Ozeri-Galai, E. et al. Failure of origin activation in response to fork stalling leads to chromosomal instability at fragile sites. Mol. Cell 43, 122–131 (2011)
Murray, J. M. & Carr, A. M. Smc5/6: a link between DNA repair and unidirectional replication? Nature Rev. Mol. Cell Biol. 9, 177–182 (2008)
Lambert, S., Watson, A., Sheedy, D. M., Martin, B. & Carr, A. M. Gross chromosomal rearrangements and elevated recombination at an inducible site-specific replication fork barrier. Cell 121, 689–702 (2005)
Mizuno, K., Lambert, S., Baldacci, G., Murray, J. M. & Carr, A. M. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 23, 2876–2886 (2009)
Lambert, S. et al. Homologous recombination restarts blocked replication forks at the expense of genome rearrangements by template exchange. Mol. Cell 39, 346–359 (2010)
Sánchez-Gorostiaga, A., Lopez-Estrano, C., Krimer, D. B., Schvartzman, J. B. & Hernandez, P. Transcription termination factor reb1p causes two replication fork barriers at its cognate sites in fission yeast ribosomal DNA in vivo. Mol. Cell. Biol. 24, 398–406 (2004)
Krings, G. & Bastia, D. swi1- and swi3-dependent and independent replication fork arrest at the ribosomal DNA of Schizosaccharomyces pombe. Proc. Natl Acad. Sci. USA 101, 14085–14090 (2004)
Williams, W. L. & Muller, U. R. Effects of palindrome size and sequence on genetic stability in the bacteriophage ϕX174 genome. J. Mol. Biol. 196, 743–755 (1987)
Voineagu, I., Narayanan, V., Lobachev, K. S. & Mirkin, S. M. Replication stalling at unstable inverted repeats: interplay between DNA hairpins and fork stabilizing proteins. Proc. Natl Acad. Sci. USA 105, 9936–9941 (2008)
Lydeard, J. R. et al. Break-induced replication requires all essential DNA replication factors except those specific for pre-RC assembly. Genes Dev. 24, 1133–1144 (2010)
Deem, A. et al. Break-induced replication is highly inaccurate. PLoS Biol. 9, e1000594 (2011)
Smith, C. E., Llorente, B. & Symington, L. S. Template switching during break-induced replication. Nature 447, 102–105 (2007)
Stephens, P. J. et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell 144, 27–40 (2011)
Zuffardi, O., Bonaglia, M., Ciccone, R. & Giorda, R. Inverted duplications deletions: underdiagnosed rearrangements? Clin. Genet. 75, 505–513 (2009)
Arlt, M. F., Wilson, T. E. & Glover, T. W. Replication stress and mechanisms of CNV formation. Curr. Opin. Genet. Dev. 22, 204–210 (2012)
Liu, P. et al. Chromosome catastrophes involve replication mechanisms generating complex genomic rearrangements. Cell 146, 889–903 (2011)
Kloosterman, W. P. et al. Constitutional chromothripsis rearrangements involve clustered double-stranded DNA breaks and nonhomologous repair mechanisms. Cell Rep. 1, 648–655 (2012)
Iraqui, I. et al. Recovery of arrested replication forks by homologous recombination is error-prone. PLoS Genet. 8, e1002976 (2012)
Pelliccia, F., Bosco, N. & Rocchi, A. Breakages at common fragile sites set boundaries of amplified regions in two leukemia cell lines K562 - Molecular characterization of FRA2H and localization of a new CFS FRA2S. Cancer Lett. 299, 37–44 (2010)
Blumrich, A. et al. The FRA2C common fragile site maps to the borders of MYCN amplicons in neuroblastoma and is associated with gross chromosomal rearrangements in different cancers. Hum. Mol. Genet. 20, 1488–1501 (2011)
Carvalho, C. M. et al. Inverted genomic segments and complex triplication rearrangements are mediated by inverted repeats in the human genome. Nature Genet. 43, 1074–1081 (2011)
Moreno, S., Klar, A. & Nurse, P. Molecular genetic analysis of fission yeast Schizosaccharomyces pombe. Methods Enzymol. 194, 795–823 (1991)
Acknowledgements
We thank E. Hoffman, J. Baxter, M. Neale and members of the Carr and Murray laboratories for discussions. J.M.M. acknowledges Cancer Research UK (CRUK) grant C9601/A9484: A.M.C. acknowledges Medical Research Council (MRC) grant G0600233.
Author information
Authors and Affiliations
Contributions
I.M., K.M. and S.A.S. performed experiments. J.M.M., K.M. and A.M.C. wrote the manuscript. All authors contributed to experimental design.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-4 and Supplementary Table 1. (PDF 464 kb)
Rights and permissions
About this article
Cite this article
Mizuno, K., Miyabe, I., Schalbetter, S. et al. Recombination-restarted replication makes inverted chromosome fusions at inverted repeats. Nature 493, 246–249 (2013). https://doi.org/10.1038/nature11676
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature11676
This article is cited by
-
Rad51-mediated replication of damaged templates relies on monoSUMOylated DDK kinase
Nature Communications (2022)
-
Protein innovation through template switching in the Saccharomyces cerevisiae lineage
Scientific Reports (2021)
-
Replication dynamics of recombination-dependent replication forks
Nature Communications (2021)
-
The nuclear pore primes recombination-dependent DNA synthesis at arrested forks by promoting SUMO removal
Nature Communications (2020)
-
Limiting homologous recombination at stalled replication forks is essential for cell viability: DNA2 to the rescue
Current Genetics (2020)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.