« Prev Next »
Phosphorylation Modification: A Universal Way to Turn on/off Signaling Pathways
Why is protein phosphorylation important? Phosphorylation is a universal protein modification in eukaryotic cells, and participates in every aspect of cellular life. The human genome encodes over 500 protein kinases that interact with diverse substrates including other kinases, transcription factors, and regulatory proteins. The cell cycle is no exception, and is tightly regulated by protein phosphorylation. Substrates are phosphorylated in a spatiotemporally specific manner by protein kinases on serine, threonine, or tyrosine residues that can subsequently be recognized by other proteins that contain a "phosphopeptide binding domain." In this way, signaling cascades initiated by protein phosphorylation trigger a series of changes in protein-protein interactions. The assembly or disassembly of protein complexes takes place at a specific time and location within the cell, leading to turning a signaling pathway "on" or "off" (Figure 1).
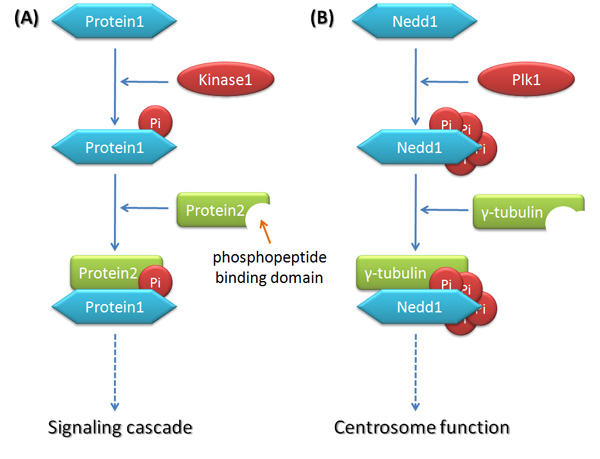
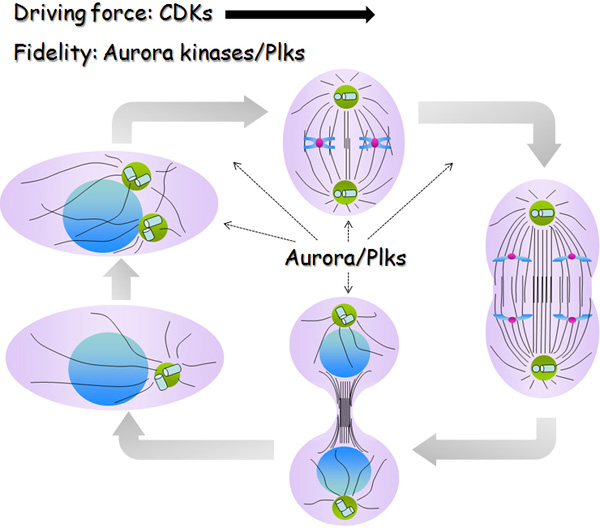
Decades ago, scientists discovered cyclin-dependent kinases (CDKs), proteins that act as master regulators of the cell cycle. As their name indicates, the activity of CDKs is controlled by cyclins, other proteins whose expression level oscillates throughout the cell cycle. Different CDKs operate during different phases of the cell cycle and provide the driving force for cell cycle progression. Besides CDKs, scientists have found several protein kinases that also control the cell cycle, primarily Plks (Polo-Like kinases) and Aurora kinases. These kinases control protein phosphorylation in a spatiotemporal manner — meaning they control where and when phosphorylation happens. In fact, these kinases are major regulators of centrosome function, spindle assembly, chromosome segregation, and cytokinesis. Unlike CDKs, which promote cell cycle progression, these kinases are responsible for avoiding errors during cell cycle progression (Figure 2; Archambault & Glover 2009, Hochegger et al. 2008, Fu et al. 2007, Barr et al. 2004, Carmena et al. 2003).
Phosphorylation Pattern: A Kinase versus a Substrate
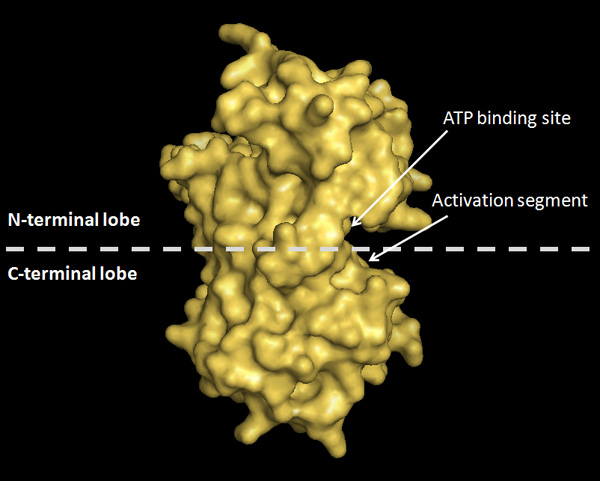
Also, we now know that the active conformations of many protein kinases are similar, and so we can classify groups of kinases based on their similar structure. For example, CDK2 and Aurora kinase possess a two-lobe architecture in their kinase domain: the N-terminal lobe and the C-terminal lobe. The active site of these kinases is buried at the interface of these two lobes, which forms a kind of groove, and also includes the ATP binding site and the kinase activation segment (Figure 3; Jeffrey et al. 1995; Fu et al. 2007).
How does the kinase-substrate interaction facilitate phosphorylation? First, the kinase has to properly position the catalytic site and ATP, and then correctly orient the substrate binding site. The kinase itself needs to be phosphorylated at a threonine residue within the activation segment. The docking portion of the substrate then fits into the groove on the surface. Then the kinase transfers a phosphate group from the ATP onto the Ser/Thr/Tyr of the substrate. Protein kinases usually get activated by binding to other entities, called activators. For example, in the absence of the activator TPX2, the activation segment of Aurora A overlaps with the substrate binding site, so that substrate binding is not optimal. With the binding of TPX2, the two lobes of Aurora A kinase domain switch to a conformation that is suitable for substrate binding and phosphoryl transfer. (Fu et al. 2007). Similarly, CDK2 also requires an interaction with Cyclin A to be fully activated (Jeffrey et al. 1995).
Collaboration between Kinases: Sequential Phosphorylation of a Single Substrate
CDK1 as a Priming Kinase to Plk1
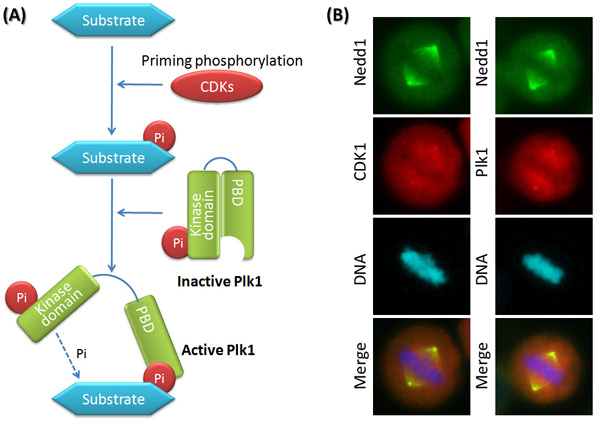
How was the collaboration among different mitotic kinases discovered? In 2003, scientists devised a proteomic screen that used phosphopeptides derived from substrates of CDKs as bait to identify proteins that were phosphorylated by CDKs. Based on the presumption that protein kinases and phosphopeptide binding domains tend to recognize overlapping sequence motifs, the screen would identify proteins/domains that would preferentially bind to the phosphopeptides that have already been primed — phosphorylated by CDKs (Elia et al. 2003). This approach led to the identification of Plk1 as a phosphopeptide binding domain, which means Plk1 coordinates with CDKs to act on the same substrates. So far, the accumulated data supports the theory that most substrates of Plk1 in metaphase are regulated by the following mechanism. The priming phosphorylation on a substrate by CDKs subsequently promotes the interaction between the substrate and Plk1. Plk1 consists of an N-terminal kinase domain and a C-terminal phosphopeptide binding domain named PBD (Polo Box Domain). In the absence of a bound substrate, the PBD forms an intramolecular interaction with the kinase domain, and inhibits the kinase activity. When a substrate is primed by another kinase like one of the CDKs, it binds to the PBD and releases the kinase domain for Plk1 function (Figure 4; Zhang et al. 2009).
Plk1 as a Priming Kinase to Aurora B
Collaboration between Kinases: One Kinase Phosphorylates Another
Aurora A Together with Bora Activates Plk1
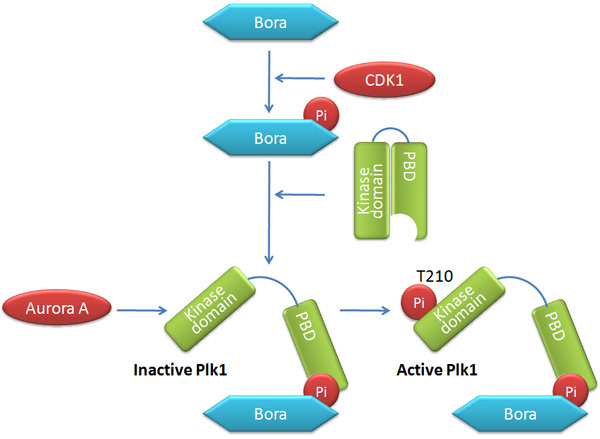
Is there another pattern of collaboration among mitotic kinases? Indeed, scientists have exposed yet another point of interaction. Besides sequential phosphorylation on single substrate, protein kinases also phosphorylate and activate each other. During the cell cycle, the activation of Plk1 relies on the phosphorylation of a threonine residue within the activation segment. Scientists recently discovered that Aurora A is responsible for activating Plk1 at the G2/M transition by phosphorylating Plk1 at the same threonine residue. Plk activation by Aurora A requires another cofactor Bora, which dramatically enhances the ability of Aurora A to phosphorylate Plk1. Interestingly, this effect of Bora is specific to Plk1, rather than all Aurora A substrates, which hints at the possibility that Bora might be working directly on Plk1. Most likely, Bora induces the conformational change of Plk1 and makes it more accessible for Aurora A, but researchers have not yet proved this. Consistent with the model above, Bora needs to be primed by phosphorylation by CDK1 before it can bind to Plk1 (Figure 5; Seki et al. 2008, Taylor et al. 2008).
Summary
References and Recommended Reading
Archambault, V. & Glover, D. M. Nature Reviews Molecular Cell Biology 10, 265–275 (2009) doi:10.1038/nrm2653.
Barr, F. A., Silljé, H. H. W. et al. Nature Reviews Molecular Cell Biology 5, 429–441 (2004) doi:10.1038/nrm1401.
Carmena, M. & Earnshaw, W. C. The cellular geography of Aurora. Nature Reviews Molecular Cell Biology 4, 842–854 (2003) doi:10.1038/nrm1245.
Fu, J., Bian, M., et al. Roles of Aurora kinases in mitosis and tumorigenesis. Molecular Cancer Research 5, 1–10 (2007) doi:10.1158/1541-7786.MCR-06-0208.
Hochegger, H., Takeda, S., et al. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nature Reviews Molecular Cell Biology 9, 910–916 (2008) doi:10.1038/nrm2510.
Jeffrey, P. D., Russo, A. A., et al. Mechanism of CDK activation revealed by the structure of a
cyclinA-CDK2 complex. Nature 376, 313–320 (1995) doi:10.1038/376313a0.
Nigg, E. A. Mitotic kinases as regulators of cell division and its checkpoints. Nature Reviews Molecular Cell Biology 2, 21–32 (2001) doi:10.1038/35048096.
Ubersax, F. A & Ferrell Jr., J. E. Mechanisms of specificity in protein phosphorylation Nature Reviews Molecular Cell Biology 8, 530–541 (2007) doi:10.1038/nrm2203.






























