« Prev Next »
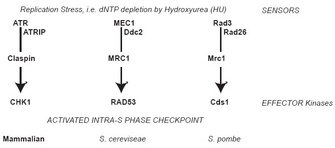
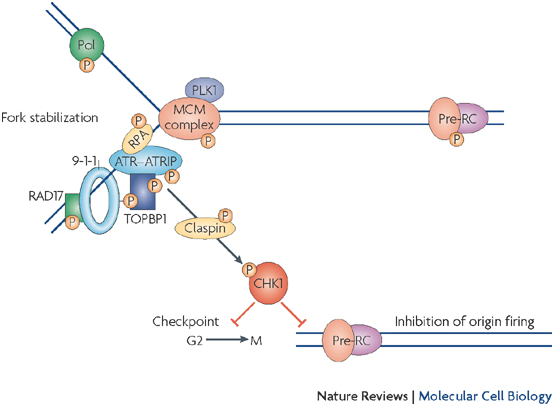
Some of the RF proteins are part of the fork protection complex (FPC), which also stabilizes the RF during HU-induced arrest. Single-stranded DNA (ssDNA) created at the stalled RF generates a signal that activates the intra-S phase checkpoint and prevents the cell cycle from moving into G2 phase until replication is complete (Figure 1). Once the replication block has been removed, either by adaptation to the drug or by the removal of HU from the system, stalled RFs resume DNA synthesis.
Although some replication origins can start in late S phase (Lopes et al. 2001), completion of replication following arrest generally depends on restart of the stalled forks (Branzei & Foiani 2007). This reliance on restarting RFs to complete whole genome synthesis raises several important questions. What signal alerts the cell that replication has paused and RF progression has stopped? How do RFs restart replication once conditions improve or the error is resolved? These questions are particularly important to human health because maintenance of genome stability during S phase helps defend against cancer (Bartkova et al. 2005).
The Unexpected Pause: Signals from Fork Stalling
Some studies in budding yeast (Saccharomyces cerevisiae) have helped us understand the function of yet another protein that participates in RF stalling: the recombination protein Rad52. This protein is recruited to the RF following HU exposure (Lisby et al. 2001). Rad52 is a homologous recombination protein required to repair DSBs, and its recruitment during HU treatment confirms two features. One is that HU exposure causes DNA damage; the other is that restarting the stalled RF involves DNA recombination. During prolonged HU exposure, the recombination and repair protein Mre11 is also recruited, again reinforcing the contention that long-term RF arrest has deleterious effects on the DNA (Lisby et al. 2004).
However, RF stalling is not a permanent event. In fission yeast (Schizosaccharyomyces pombe) the Rpa signal at a stalled RF is lost over time, while the Rad52 homolog Rad22 is recruited (Irmisch et al. 2009). Rad52/Rad22 and Rad51 recombination proteins are known to displace Rpa during homologous recombination (Kurokawa et al. 2008; Seong et al. 2009; Sugiyama & Kantake 2009). So it appears that the ssDNA-Rpa signal times out and switches to ssDNA-Rad51 in order to promote recombination-dependent RF restart mechanisms. Is the HU-induced DNA damage reversible or permanent? At the level of single molecules, it appears the answer is that DNA damage persists. Single-molecule "DNA fiber" analysis demonstrated that DNA damage signals increase with extended HU treatment, and the damage signal persists even after HU is removed from the system (Petermann et al. 2010). It is clear that the replication checkpoint is linked to the DNA damage checkpoint, and DNA damage triggers the checkpoint signal that prohibits progression to metaphase (Figure 3). Perhaps the accumulation of damage and the inability to restart with prolonged HU exposure are due to the loss of components from stalled forks. Could a switch from Rpa to Rad51 be a signal to start repair and consequently restart the stalled RF?
Recovery and Restart of DNA Replication
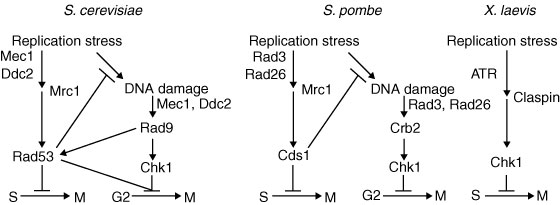
Once the block to replication is resolved, what signals the restarting of replication? One possible mechanism is protein phosphorylation, since several RF components are phosphorylated upon stalling (Figure 4). Mrc1, which is part of the FPC, is phosphorylated during fork stalling and then interacts with downstream effector kinases to transmit the checkpoint signal (see Figure 1; Bjergbaek et al. 2005; Lou et al. 2008; Xu et al. 2006). These protein modifications not only activate checkpoint response but can also attenuate the checkpoint, and Mrc1 is once again a good example. The Xenopus homolog of Mrc1, Claspin, is also phosphorylated by polo-like kinase, Plx1, causing Claspin to fall off of chromatin and stop the checkpoint signal (Yoo et al. 2004). In S. cerevisiae, Mrc1 phosphorylation changes its association with N- and C-terminal domains of the polymerase, Polε, which may modify polymerase association with the RF and its function (Lou et al. 2008). Are phosphorylated RF components simply modified, or are they lost and replaced at the fork? Does RF protein dephosphorylation reactivate the RF?
The recombinase proteins Rad51 and Rad52 move to the nucleus with prolonged HU exposure and during restart, again emphasizing the essential role of recombination for restarting the fork (Hanada et al. 2007; Irmisch et al. 2009; Petermann et al. 2010; Torres et al. 2004; Wray et al. 2008). Corroborating evidence for this observation is that RAD51 mutant yeast cells (∆rhp51 in S. pombe) show reduced RF recovery and mitotic defects (Bailis et al. 2008). However, too much unchecked recombination can prevent fork restart (Bernstein et al. 2009; Liberi et al. 2005; Shimura et al. 2008; Willis & Rhind 2009; Yodh et al. 2009). Is there an intrinsic feature of the stalled RF that requires recombination activity to allow restart?
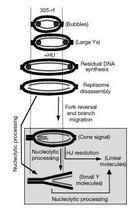
Checkpoint resolution and fork recovery depend on the creation of some ssDNA, as well as DNA damage repair, to restart replication. In addition, DNA nucleases help resolve stalled structures and are regulated by S phase checkpoint effector kinases: Chk1 in metazoa; Rad53 in Saccharomyces cerevisiae; Cds1 in Schizosaccharomyces pombe (Figure 1). Loss of these kinases results in excessive DNA unwinding in the presence of HU, producing an extended amount of ssDNA that becomes coated with RPA (Feng et al. 2006; Lucca et al. 2004; MacDougall et al. 2007; Sogo et al. 2002) and likely reflects uncoupling of the MCM helicase from the RF. Furthermore, the absence of effector kinases causes DNA damage with extensive formation of DSBs, which are likely caused by unresolvable DNA structures (Figure 5; Bailis et al, 2008; Feng et al. 2006; Lindsay et al. 1998; Lucca et al. 2004; Marchetti et al. 2002; Sogo et al. 2002).
Summary
The FPC stabilizes the RF during stalling. Protein modifications (e.g., phosphorylation) set up a signal cascade to promote stalling and maintain the fork structure. RF stalling must occur regularly during genome duplication, even in the absence of external DNA-damaging agents, resulting simply from sequence repetition or collision with DNA metabolism enzymes (Baird 2008; Lee et al. 2007; Mirkin & Mirkin 2007; Samadashwily et al. 1997). The stalling signal is deactivated when cells enter conditions allowing fork restart after DNA damage has been resolved. In addition, recombination and repair proteins are involved in replication restart, creating and repairing DNA breaks so that the fork can resume synthesis later. Research in yeast and metazoan systems to understand the nature of the stalled fork and restart due to HU is advancing our understanding of the processes involved, as well as the problems that develop when components are missing. The interplay between checkpoint signaling and break induction/recombination regulates HU response and stability in treated cells. Above, we discussed several detailed mechanisms for managing the quality of DNA replication, and the implications of these findings have a major impact on our understanding of how cells prevent carcinogenesis (Bartkova et al. 2005).
References and Recommended Reading
Alcasabas, A. A. et al. Mrc1 transduces signals of DNA replication stress to activate Rad53. Nature Cell Biology 3, 958–965 (2001)
Bailis, J. M. et al. Minichromosome maintenance proteins interact with checkpoint and recombination proteins to promote s-phase genome stability. Molecular and Cellular Biology 28, 1724–1738 (2008)
Baird, D. M. Telomere dynamics in human cells. Biochimie 90, 116–121 (2008)
Bartkova, J. et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature 434, 864–870 (2005)
Bernstein, K. A. et al. Sgs1 function in the repair of DNA replication intermediates is separable from its role in homologous recombinational repair. EMBO Journal 28, 915–925 (2009)
Bjergbaek, L. et al. Mechanistically distinct roles for Sgs1p in checkpoint activation and replication fork maintenance. EMBO Journal 24, 405–417 (2005)
Branzei, D. & Foiani, M. Interplay of replication checkpoints and repair proteins at stalled replication forks. DNA Repair (Amsterdam) 6, 994–1003 (2007)
Branzei, D. & Foiani, M. The Rad53 signal transduction pathway: Replication fork stabilization, DNA repair, and adaptation. Experimental Cell Research 312, 2654–2659 (2006)
Cimprich, K. A. & Cortez, D. ATR: An essential regulator of genome integrity. Nature Reviews Molecular and Cellular Biology 9, 616–627 (2008)
Feng, W. et al. Genomic mapping of single-stranded DNA in hydroxyurea-challenged yeasts identifies origins of replication. Nature Cell Biology 8, 148–155 (2006)
Froget, B. et al. Cleavage of stalled forks by fission yeast Mus81/Eme1 in absence of DNA replication checkpoint. Molecular Biology of the Cell 19, 445–456 (2008)
Hanada, K. et al. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nature Structural & Molecular Biology 14, 1096–1104 (2007)
Hiyama, T. et al. Haploinsufficiency of the Mus81-Eme1 endonuclease activates the intra-S-phase and G2/M checkpoints and promotes rereplication in human cells. Nucleic Acids Research 34, 880–892 (2006)
Irmisch, A. et al. Smc5/6 maintains stalled replication forks in a recombination-competent conformation. EMBO Journal 28, 144–155 (2009)
Kai, M. et al. Replication checkpoint kinase Cds1 regulates Mus81 to preserve genome integrity during replication stress. Genes & Development 19, 919–932 (2005)
Kanoh, Y., Tamai, K. & Shirahige, K. Different requirements for the association of ATR-ATRIP and 9-1-1 to the stalled replication forks. Gene 377, 88–95 (2006)
Kemp, M. & Sancar, A. DNA distress: Just ring 9-1-1. Current Biology 19, R733–R734 (2009)
Kurokawa, Y. et al. Reconstitution of DNA strand exchange mediated by Rhp51 recombinase and two mediators. PLoS Biology 6, e88 (2008)
Lee, J. A., Carvalho, C. M. & Lupski, J. R. A DNA replication mechanism for generating nonrecurrent rearrangements associated with genomic disorders. Cell 131, 1235–1247 (2007)
Liberi, G. et al. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes & Development 19, 339–350 (2005)
Lindsay, H. D. et al. S-phase-specific activation of Cds1 kinase defines a subpathway of the checkpoint response in Schizosaccharomyces pombe. Genes & Development 12, 382–395 (1998)
Lisby, M., Rothstein, R. & Mortensen, U. H. Rad52 forms DNA repair and recombination centers during S phase. PNAS 98, 8276–8282 (2001)
Lisby, M. et al. Choreography of the DNA damage response: Spatiotemporal relationships among checkpoint and repair proteins. Cell 118, 699–713 (2004)
Lopes, M. et al. The DNA replication checkpoint response stabilizes stalled replication forks. Nature 412, 557–561 (2001)
Lou, H. et al. Mrc1 and DNA polymerase epsilon function together in linking DNA replication and the S phase checkpoint. Molecular Cell 32, 106–117 (2008)
Lucca, C. et al. Checkpoint-mediated control of replisome-fork association and signalling in response to replication pausing. Oncogene 23, 1206–1213 (2004)
MacDougall, C. A. et al. The structural determinants of checkpoint activation. Genes & Development 21, 898–903 (2007)
Marchetti, M. A. et al. A single unbranched S-phase DNA damage and replication fork blockage checkpoint pathway. PNAS 99, 7472–7477 (2002)
Mirkin, E. V. & Mirkin, S. M. Replication fork stalling at natural impediments. Microbiology and Molecular Biology Reviews 71, 13–35 (2007)
Petermann, E. et al. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Molecular Cell 37, 492–502 (2010)
Samadashwily, G. M., Raca, G. & Mirkin, S. M. Trinucleotide repeats affect DNA replication in vivo. Nature Genetics 17, 298–304 (1997)
Seong, C. et al. Regulation of Rad51 recombinase presynaptic filament assembly via interactions with the Rad52 mediator and the Srs2 anti-recombinase. Journal of Biological Chemistry 284, 24363–24371 (2009)
Shimura, T. et al. Bloom's syndrome helicase and Mus81 are required to induce transient double-strand DNA breaks in response to DNA replication stress. Journal of Molecular Biology 375, 1152–1164 (2008)
Smith, K. D., Fu, M. A. & Brown, E. J. Tim-Tipin dysfunction creates an indispensible reliance on the ATR-Chk1 pathway for continued DNA synthesis. Journal of Cell Biology 187, 15–23 (2009)
Sogo, J. M., Lopes, M. & Foiani, M. Fork reversal and ssDNA accumulation at stalled replication forks owing to checkpoint defects. Science 297, 599–602 (2002)
Sugiyama, T. & Kantake, N. Dynamic regulatory interactions of rad51, rad52, and replication protein-a in recombination intermediates. Journal of Molecular Biology 390, 45–55 (2009)
Torres, J. Z., Schnakenberg, S. L. & Zakian, V. A. Saccharomyces cerevisiae Rrm3p DNA helicase promotes genome integrity by preventing replication fork stalling: Viability of rrm3 cells requires the intra-S-phase checkpoint and fork restart activities. Molecular and Cellular Biology 24, 3198–3212 (2004)
Van, C. et al. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. Journal of Cell Biology 189, 233–246 (2010)
Willis, N. & Rhind, N. Mus81, Rhp51(Rad51), and Rqh1 form an epistatic pathway required for the S-phase DNA damage checkpoint. Molecular Biology of the Cell 20, 819–833 (2009)
Wray, J. et al. Distinct RAD51 associations with RAD52 and BCCIP in response to DNA damage and replication stress. Cancer Research 68, 2699–2707 (2008)
Xu, Y. J., Davenport, M. & Kelly, T. J. Two-stage mechanism for activation of the DNA replication checkpoint kinase Cds1 in fission yeast. Genes & Development 20, 990–1003 (2006)
Yan, S. & Michael, W. M. TopBP1 and DNA polymerase-alpha directly recruit the 9-1-1 complex to stalled DNA replication forks. Journal of Cell Biology 184, 793–804 (2009a)
Yan, S. & Michael, W. M. TopBP1 and DNA polymerase alpha-mediated recruitment of the 9-1-1 complex to stalled replication forks: Implications for a replication restart-based mechanism for ATR checkpoint activation. Cell Cycle 8, 2877–2884 (2009b)
Yodh, J. G. et al. BLM helicase measures DNA unwound before switching strands and hRPA promotes unwinding reinitiation. EMBO Journal 28, 405–416 (2009)
Yoo, H. Y. et al. Adaptation of a DNA replication checkpoint response depends upon inactivation of Claspin by the Polo-like kinase. Cell 117, 575–588 (2004)
Zachos, G., Rainey, M. D. & Gillespie, D. A. Chk1-dependent S-M checkpoint delay in vertebrate cells is linked to maintenance of viable replication structures. Molecular and Cellular Biology 25, 563–574 (2005)
Zou, L., Liu, D. & Elledge, S. J. Replication protein A-mediated recruitment and activation of Rad17 complexes. PNAS 100, 13827–13832 (2003)



 Figure 1: Intra-S phase checkpoint signaling
Figure 1: Intra-S phase checkpoint signaling