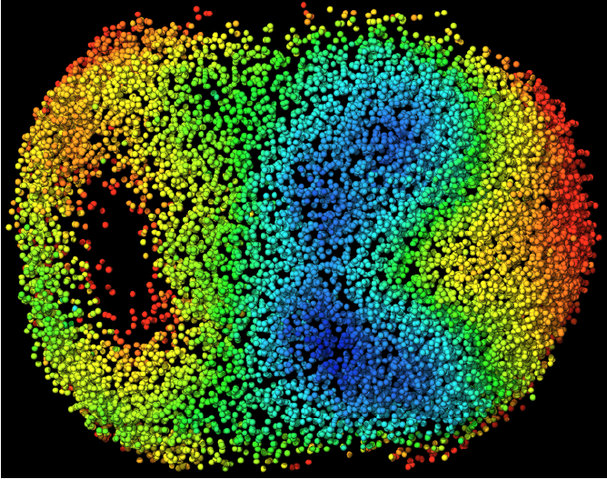
Visual model of cellular motion in a developing mouse embryo. Cells are color coded by their average speed of movement.
Credit: Reprinted from Cell, Vol 175, Issue 3, McDole, K. et al. P859-876.E33, Copyright 2018, with permission from Elsevier.
Stop, Start, Rewind: A front-seat view of mouse development
A new microscope allows researchers to track single cells in developing embryos, forward and backward in time.
20 December 2018
Live imaging of mouse development from gastrulation to early organogenesis.
The video begins with a small electric-blue sphere that shimmers in the dark. The ball pulses and grows, the uniform blue resolving into points as the object — now more of an amorphous blob than a sphere — expands to fill the screen.
A delicate choreography becomes apparent as the glittering speckles, each one a fluorescently labelled cell, begin to move in synchrony. As the video comes to an end, a void opens to the left, the first hint of the tissue that will become the animal’s gut. A counter marks the time: 43:30:00 — 43.5 hours compressed into 26 seconds.
The video captures a critical phase in mouse embryogenesis, days 6.5 to 8.5 of development, as the embryo begins its transformation from unremarkable ball of cells to burgeoning vertebrate. It was taken using a microscope developed by physicist, Philipp Keller, and his team at the Howard Hughes Medical Institute’s Janelia Research Campus in Ashburn, Virginia.
Reprinted from Cell, Vol 175, Issue 3, McDole, K. et al. P859-876.E33, Copyright 2018, with permission from Elsevier.
The first light-sheet microscope for observing mouse embryo development at the single-cell level.According to Keller, the video — accompanying a paper published in Cell in October 2018 — represents the first time such a sequence has been observed, at least over such a lengthy period and with such high resolution. (The microscope was able to capture the entire embryo, of 50,000 cells, every 3 to 5 minutes for nearly two days.) Previously, Keller explains, researchers wishing to view such processes had to make sacrifices, of spatial resolution, temporal resolution, field of view, or total experiment time.
Keller and his team have been chipping away at those limitations for at least a decade, thanks to a technology called light-sheet microscopy. The latest iteration, he says, represents their best design yet. For the researchers now working with Keller to exploit the technology, the microscope is proving transformative, facilitating experiments not previously possible.
Keller emphasizes that the microscope can be used to study any tissue or organism, or even artificial three-dimensional cell structures called organoids. Researchers can access the hardware via collaboration, or by requesting time at the Janelia Advanced Imaging Center, which makes the hardware available at no cost. Keller estimates the microscope would cost US$400,000 to US$500,000 to build.
In plane view
In a light-sheet microscope, the sample is effectively sliced with a razor-thin plane (or sheet) of light, inducing fluorescence in a tight band. That signal is imaged using a camera positioned perpendicular to the sheet. By moving the sheet up or down through the sample, the instrument captures a stack of images that can be computationally reconstructed into a 3D volume. Fast and requiring relatively low-intensity light, the process is gentle on sensitive biological samples, allowing for exceptionally long-term imaging.
Keller’s first instrument, the DLSM (digital scanned light-sheet microscope) was comparatively simple, including just two ‘arms’ for illumination and detection. Subsequent iterations have added a second pair of arms and another camera to penetrate deeper samples, as well as adaptive imaging to accommodate the optical distortions that occur as light penetrates thick biological tissue. With each new version, the team advanced to ever more complex specimens, from transparent zebrafish, to opaque fruit flies. But they had never been able to capture a developing mouse.
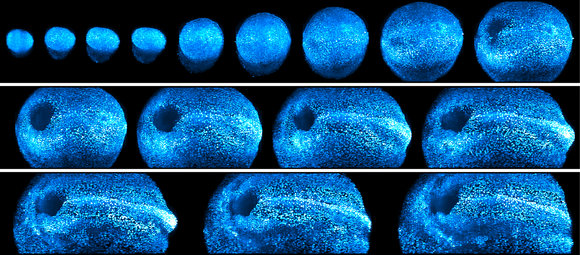
Reprinted from Cell, Vol 175, Issue 3, McDole, K. et al. P859-876.E33, Copyright 2018, with permission from Elsevier.
Snapshots of a developing mouse embryo over 44 hours of continuous imaging.Mouse embryogenesis represents “the broadest and most difficult set of challenges” his lab has yet encountered, Keller says. Most obviously, unlike animals that develop in eggs, mouse embryos normally grow in the absence of light. “The mouse embryo really can’t handle light very well,” he says.
A growing embryo also requires tightly controlled environmental conditions such as temperature and pH, as well as the freedom to move and grow. As a result, when earlier researchers tried to image embryos for extended periods, the samples tended to drift in and out of focus and out of frame.
Led by postdoctoral researcher, Kate McDole, Keller and his team built a system to overcome this propensity, incorporating and advancing on the elements they had been perfecting for years. The microscope features four custom arms, two each for illumination and detection, designed to peer through the murky growth media that embryos favour. An environmental chamber supports growth, while an adaptive imaging system tracks the specimen over time, tweaking the light paths to accommodate the sample’s ever-changing optical properties. Because the system can use low-intensity lighting to capture its images, samples can be imaged for days without apparent ‘phototoxicity’.
To analyze the resulting data, the research team developed a suite of open-source software to locate cells, track where they go, identify cell divisions, and compare and register those behaviours across individuals — and to do so automatically. The microscope generates so much data, Keller explains, that manually tracing every cell through one 48-hour dataset would require two to three years of work. “We have probably on the order of about 100 of these datasets. So we’re talking about hundreds of years of work to do this in a statistically meaningful way.”
The software can perform the same task in days. Users can select any arbitrary cell in a time-lapse dataset, say, the first indicators of a developing organ, and play development backward or forward to see where it came from or where it goes, and how that behaviour differs across samples.
Tracing cells
Sonja Nowotschin, a senior research scientist at the Memorial Sloan Kettering Cancer Center in New York, studies the development of the early endoderm during mouse development, which gives rise to the respiratory and digestive systems. She is collaborating with Keller, and plans to use the new microscope at Janelia in early 2019.
Nowotschin says the new microscope design is gentle enough for her to trace cellular paths that were previously difficult to detect. In live-cell microscopy, she explains, researchers can rarely have it all: they must sacrifice either time, resolution, or depth, or risk damaging the sample.
With the new microscope, “we can look at complex cell behaviours that we couldn’t do before at a single-cell level.”
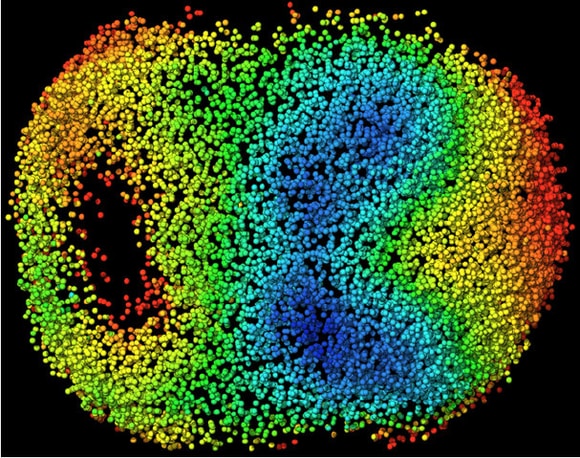
Reprinted from Cell, Vol 175, Issue 3, McDole, K. et al. P859-876.E33, Copyright 2018, with permission from Elsevier.
Visual model of cellular motion in a developing mouse embryo. Cells are color coded by their average speed of movement.Keller and his team have used their own data to build a database of cellular motion that has been averaged across multiple specimens — a first stab at identifying the ‘stereotypical’ behaviour of development. Their analysis of that dataset revealed a surprise, he says.
The team probed the orientation of structures associated with cellular division, called mitotic spindles, throughout the embryo. They found that as the embryo elongates, folds, and differentiates, mitotic spindles do not orient randomly; instead, they mirror the physical forces acting on the tissue itself. As the primitive nervous system elongates, for instance, mitotic spindles tend to align parallel to that elongation axis.
“Nobody has seen that before,” he explains. The question now, is one of causality. “Is it the cell divisions that make it possible for the tissue to fold up like that, to elongate like that? Or is it the other way around, that because we have these dramatic forces acting on these tissues, they force all of the cell divisions in a particular orientation?”
Shankar Srinivas, a developmental biologist at the University of Oxford, UK, has used the microscope in collaboration with Keller. Srinivas studies cardiac development in the mouse. In his lab, he has a confocal microscope and a commercial light-sheet microscope. But neither is capable of imaging the developing embryo with sufficient sensitivity and resolution for the experiments he wants to do.
Working with Keller’s team, Srinivas and postdoc, Richard Tyser, studied the contribution of a particular cellular population — a handful of cells found at the convergence of different tissues — to the developing heart.
Early in embryogenesis, these cells keep one particular gene off — the mammalian version of a fly gene called tinman, named for the character in The Wizard of Oz who lacks a heart. But as they migrate through the embryo, the team found in as-yet-unpublished work, these cells turn on that gene, confirming its role in the heart. It’s a finding they could only work out by physically tracking where those cells actually go as the embryo develops. “That’s where the collaboration with Philipp was so valuable,” says Srinivas.
