
Credit: Illustration by Sam Falconer
Canada’s scientists are elucidating the dark metabolome
Teams studying the human body at the molecular level are grappling with matter that defies identification.
19 November 2021

Illustration by Sam Falconer
When biochemists analyse the metabolome — the entire set of small molecules in a cell or tissue sample — they would be lucky to identify a mere fraction of them. In advanced analytical-chemistry experiments, the signature produced by the vast majority of these molecules, called metabolites, including peptides, lipids and carbohydrates, will not match any known molecular structure.
“It’s a puzzle that keeps you up at night,” says Rafael Montenegro-Burke, who relocated from the United States to Canada in 2020 to study the unidentified majority of the human metabolome, known as the ‘dark metabolome’, as part of his work at the University of Toronto’s Donnelly Centre for Cellular and Biomolecular Research, in Toronto, Canada.
A reference to dark matter, the elusive energy and matter components of the Universe, the dark metabolome describes “the stuff we keep on seeing, using instruments like mass spectrometers, but which we just can’t identify”, says David Wishart, a metabolomics researcher at the University of Alberta in Edmonton, Canada.
We run a sample, then look through all of the known compounds, and 95% of the signal we still can’t identify.” If we don’t know what the dark metabolome is, says Wishart, we can’t know how important it is, particularly in the context of human health.
When the field of metabolomics emerged about 20 years ago, the whole human metabolome was essentially dark matter, says Wishart. “Around 2005, I started a project called the Human Metabolome Project, comparable to the Human Genome Project, to try to identify all known compounds in the human body,” he says.
The aim was to create a freely accessible database of human metabolites. At the time, the list of unknown compounds totalled around 2,180, says Wishart. It’s since ballooned to more than 170,000. “We think it will probably be more than 4 million by the time we are done,” he says.
Metabolomics is the newest of the ’omics research fields, which investigate cells, tissues or whole organisms from a cascading set of vantage points. Genomics takes the highest-level approach, mapping the entire set of genes in the organism’s DNA; transcriptomics looks at the messenger RNA (mRNA), copied from the DNA.
Proteomics is the study of proteins, which result from mRNA translation. Drilling all the way down, metabolomics looks at the small molecules, defined as being less than 1,500 daltons in size (one dalton is about the mass of one hydrogen atom) that result from cellular metabolism.
They are influenced not only by our genes and proteins, but also by what we’ve eaten, breathed in or otherwise been exposed to, as well as the complex array of metabolites produced by our gut microbiome.
Montenegro-Burke is mapping the dark metabolome of a range of organisms relevant to human-health research, such as mice, yeast, fruit flies and roundworms. He uses mass spectrometry to measure the molecular weight of each unknown metabolite, and to assess the characteristic way each compound breaks into fragments during analysis, which generates a fingerprint rich in structural information.
The metabolome, which varies minute-by-minute in response to internal and external stimuli, informs research on the connection between the genetic make-up of a cell or organism (the genotype) and its observable traits (the phenotype). But, securing funding for metabolomics research is difficult. Although most well-funded universities have a shared-access mass spectrometer, metabolomics researchers require their own high-end machine.
“Without a high-throughput mass spectrometer, you cannot do metabolomics,” says Sara Sharifpoor, director of research operations and strategy at the Donnelly. Montenegro-Burke was “very lucky” to receive a Can$400,000 (US$313,266) Canadian Foundation for Innovation infrastructure grant in August, she says.
“Because the up-front investment is high, and it takes a long time to generate the data, in the Canadian funding climate, it’s harder to make the case to say that this work is immediately relevant.”
Dark-metabolome research may be eligible for Natural Sciences and Engineering Research Council of Canada (NSERC) funding, but “is so resource-intensive, there’s no way you could do it on an NSERC grant”, says Ian Lewis, a researcher at the University of Calgary in Alberta, Canada.
Lewis was part of a host-microbiome study that showed how a common antirejection drug caused colitis as a side effect. The metabolized drug was being reactivated in the intestine by a previously unrecognized enzyme released by a gut bacterium (M. R. Taylor et al. Sci. Adv. 5, eaax2358; 2019).
Mapping the dark metabolome calls for expertise in biology, chemistry and analytical techniques.
“It also requires a good understanding of cheminformatics, machine learning and artificial intelligence,” says Wishart. That means assembling a multidisciplinary team. “At the University of Alberta, there’s myself in biology, then a group led by Liang Li in chemistry, and a group in machine learning and computing science led by Russ Greiner.”
Metabolomics collaborations extend across Canada. Lewis and Wishart are two of eight ‘node leaders’ of a Canada-wide network, the Metabolomics Innovation Centre, set up as a joint initiative between the University of Alberta and the University of Victoria in 2011. Monthly discussions help to consolidate collaboration across the network, which now encompasses researchers across five universities.
With the push into dark metabolomics over the past decade, there’s not much more that can be done with standard mass spectrometry and nuclear magnetic resonance spectroscopy (NMR) techniques, Lewis notes. NMR, a complementary technique, identifies molecules by measuring the characteristic response of their component atoms to a magnetic field.
“This is now being driven by people at the edge of the technology bubble,” says Lewis. “The very best mass-spectrometry and NMR people are the ones who are really pushing that boundary.”
As the number of unknown compounds in the human metabolome has grown, so too has the focus of the University of Alberta team on developing computational tools to help identify more of the mystery compounds detected by mass spectrometry and NMR.
“We have moved on from trying to enumerate these compounds by hand, using instrumentation, to asking what computers can do to help us with the dark metabolome,” says Wishart. The team is developing software to predict which molecular structures might conceivably be found in a sample, and what the mass spectra of the expected metabolites could look like.
Computational approaches cannot routinely and unambiguously assign the structure of most mystery molecules in a sample, based on mass spectrometry. Increasingly, however, the software can at least identify the class of organic compound that they belong to.
"That’s still a big win,” says Wishart. From identifying only 5% of the human metabolome, the team can now assign the compound class of up to half of the molecules in a sample, he says, which is benefiting other areas of research.
“[Using] the software we developed to help us predict mass spectra, we have been working with forensic chemists to help them identify new street drugs,” says Wishart. The same tools could also help accelerate the search for natural products with potentially useful properties.
“It’s that analogy of why go to the Moon,” says Wishart. “The process of getting there allowed us to develop computers and rocket-engine technology, and the concept of team science. The dark metabolome has created a whole raft of practical solutions that are rippling through all of chemistry, hopefully helping us to understand the chemical universe around us.”
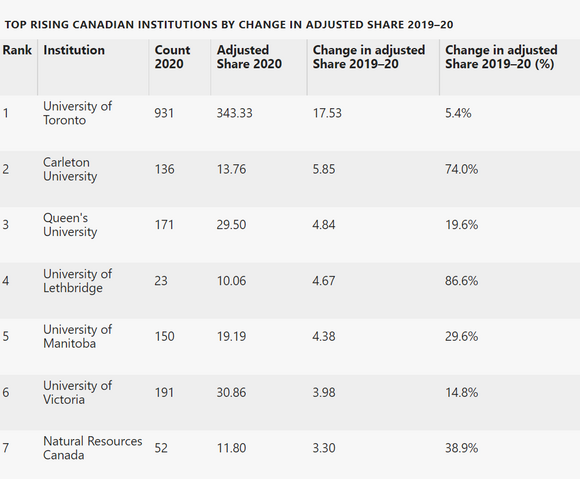
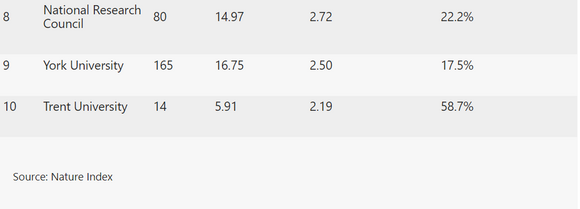
This article is part of Nature Index Canada, an editorially independent supplement. Advertisers have no influence over the content.
