Abstract
B7 family members act as co-stimulatory or co-inhibitory molecules in the adaptive immune system. Thisstudy aimed to investigate the dysregulation, prognostic value and regulatory network of B7 family members in non-small cell lung cancer (NSCLC). Data for lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) patients were extracted from public databases. Patient prognosis was determined by Kaplan–Meier analysis. The downstream signaling pathways of B7 family were identified via GO and KEGG analysis. The key B7 related genes were selected by network, correlation and functional annotation analysis. Most B7 family members were dysregulated in LUAD and LUSC. The expression of B7-1/2/H3 and B7-H5 were significantly associated with overall survival in LUAD and LUSC, respectively. The major pathway affected by B7 family was the EGFR tyrosine kinase inhibitor resistance and ErbB signaling pathway. MAPK1, MAPK3 and MAP2K1 were pivotal B7 related genes in both LUAD and LUSC. This study reveals an overall dysregulation of B7 family members in NSCLC and highlights the potential of combination use of tyrosine kinase inhibitors or MEK/ERK inhibitors with B7 member blockade for NSCLC treatment.
Similar content being viewed by others
Introduction
Lung cancer is the most common cancer type with 2.1 million new cases diagnosed in 2018, accounting for 12% of newly diagnosed cancer cases1. It is also the first cause of worldwide cancer death with estimated 1.6 million deaths annually that accounted for 20% of total cancer-related death with very low 5-year survival rate1,2,3. According to the IARC report on cancer incidence and mortality in 2020: 2,206,771 new cases (about 11.4% of all cancer cases) and 1,796,144 new deaths of lung cancer (about 18.0% of cancer deaths) in 2020. Lung cancer is the leading cause of cancer death and is also the most common type of cancer in men and the second most common type of cancer in women4. Non-small cell lung cancer (NSCLC) accounts for approximately 85% of lung cancer5. Lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) are the two most common subtypes of NSCLC, accounting for 65–70% of all lung cancers6. Smoking is currently considered to be the most important risk factor for lung cancer7,8,9. A majority of patients are diagnosed with advanced or metastatic disease without apparent symptoms10,11. Currently, there are limited treatment methods for NSCLC, with chemotherapy being the main clinical option2 In recent years, tumor immunotherapy has been applied in some cases in NSCLC patients and the advancement of it has improved patient survival and reduced suffering in some cancer patients, providing important information for diagnosis and treatment of NSCLC6,12.
The B7 family is defined as co-stimulating or co-inhibiting molecules in the activation of T cells and their ligands and receptors play critical roles in the adaptive immune response and malignant tumors13. It belongs to IgSF (immunoglobulin super-family) and its outer membrane domain contains a different amount of immunoglobulin domain14. In the past decades, the B7 family has been found to include 10 members: B7-1(CD80), B7-2(CD86), B7-DC(PD-L2 or CD273), B7-H1(PD-L1 or CD274), B7-H2(ICOSLG), B7-H3(CD276), B7-H4(VTCN1), B7-H5(VISTA), B7-H6(NCR3LG1) and B7-H7(HHLA2)15. Studies have shown that many inhibitory B7 family members are highly expressed in tumors and participate in tumor cell immunosuppression through a negative second signal16,17,18. Abundant inhibitory signals may weaken the response of T cells19. Experimental evidence suggests that manipulation of the B7 family may affect anti-tumor immunity, thus the B7 family and its receptors hold great potential as targets for tumor immune checkpoint therapy20.
To date, the pathogenesis of NSCLC, including the epigenetic regulation of mRNA and protein changes has not yet been fully elucidated, and the molecular mechanism needs to be further explored to make certain about the progress of therapeutic options for the occurrence and development of NSCLC. Strikingly, notwithstanding these open questions, increasing studies have suggested that the introduction of targeted therapy and immune checkpoint inhibitors has greatly improved the treatment of patients with NSCLC compared with traditional therapeutic approaches8,9. Some genes also contribute to dissimilar degrees of cell invasion in NSCLC21. Therefore, formulating related agents for certain immune checkpoint inhibitors to effectively inhibit the occurrence and development of NSCLC cells has become another mainstream way22. In this study, based on the public data, we analyzed the mRNA expression level, gene alterations, DNA methylation, prognostic value, functional network of the B7 family and identified the B7 family regulatory network in NSCLC. The findings may help better understand the molecular mechanism of NSCLC and provide information on favorable combined immunotherapy of NSCLC patients in the future.
Results
Dysregulation of B7 family members in NSCLC
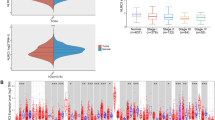
Heatmap showing the expression levels of B7 family member in LUAD and LUSC was constructed using TCGA data (Fig. 1a). Overall, the expression levels of B7-H3 and B7-H5 were relatively higher while the expression level of B7-H6 was lower than that of other B7 members in both LUAD and LUSC. TCGA and GTEx data were then combined to compare the expression levels of B7 family member between normal and tumor tissues in LUAD and LUSC (Fig. 1b). The results showed that B7-1, B7-2, B7-DC, B7-H1, B7-H2, B7-H5, B7-H6 were consistently down-regulated in both LUAD and LUSC, but B7-H3 and B7-H4 were up-regulated. Particularly, the level of B7-H7 was increased in LUAD but decreased in LUSC. To unveil the dysregulation mechanism of B7 family members in NSCLC, we investigated the relationship between the expression of B7 family members and DNA methylation and copy number alteration, respectively (Fig. 1c, d). The mRNA expression of B7-1/2/DC/H3/H4/H5/H7 were inversely correlated with DNA methylation level in LUAD and the expression of B7-H1 and B7-H4 were also negatively associated with DNA methylation in LUSC (Fig. 1c), suggesting that the dysregulation of B7 members might be partially due to promotor DNA hyper/hypo-methylation in NSCLC. Moreover, the expression level of B7-1, B7-DC, B7-H1, B7-H2, B7-H3, B7-H4, B7-H5, B7-H6 were significantly associated with gene amplification, diploid and copy number increase in LUAD. In the meantime, the expression of B7-1, B7-2, B7-DC, B7-H1, B7-H2, B7-H3, B7-H4, B7-H5, B7-H6 were significantly associated with copy number status in LUSC (Fig. 1d). Collectively, our data suggests that both DNA methylation and copy number alteration may be involved in the dysregulation of B7 family members in NSCLC.
Altered expression and underlying mechanism of B7 family in different subtypes of NSCLC. (a) The expression profile of B7 family was displayed in heatmap in normal and tumor tissues of different NSCLC patients with data extracted from TCGA database. (b) The violin plots showed the average expression level of B7 family members in normal and tumor tissues based on TCGA and GTEX data. Expression level is mapped by Transcripts Per Kilobase of exon model per Million mapped reads (TPM). (LUAD:512 tumor samples and 347 normal samples; LUSC:498 tumor samples and 336 normal samples). (c) Correlation between promoter DNA methylation and the expression of B7 family. (d) The relationship between expression and copy numberstatus of B7 family members (*P < 0.05; **P < 0.01; ***P < 0.001).
Genetic alterations of B7 family members in NSCLC
To further understand the dysregulation of B7 members in NSCLC, we also determined their mutations. The frequency of B7 member mutation, amplification and deep deletion in LUAD and LUSC was shown in Fig. 2a. Overall, the mutation rates of B7 members were relatively higher in LUSC than LUAD. B7-1, B7-2 and B7-H7 exhibited higher mutation rates than other B7 members in LUSC and the major mutation type for them and B7-H5, B7-H6 was amplification. The major mutation type found for B7-DC, B7-H1 and B7-H4 was deep deletion. At the same time, Fig. 2b demonstrated the total mutation rate and different mutation types of B7 members. Similar with the results in Fig. 2a, the mutation rates for B7-DC, B7-H1 and B7-H4 were relatively higher in both LUAD (4%, 4% and 2.9%) and LUSC (5%, 5% and 4%). But the mutation rates were even higher for B7-1 (9%), B7-2 (10%) and B7-H7 (9%) in LUSC but not LUAD.
Genetic alterations of B7 family in different subtypes of NSCLC. (a) The frequency of genetic alterations in B7 family members in NSCLC. The types of changes include mutations (green), amplifications (red), and deep deletions (blue). (b) The gene mutation rate of B7 family members was analyzed, including missense mutations (green), truncation mutations (dark gray), amplification (red), deep deletion (blue) and no change (gray).
Association of B7 family member expression with clinicopathological parameters and survival
We further assessed the association between B7 family member expression and some clinicopathological parameters including cancer status (with tumor, tumor free), gender (male, female) and pathological stage (stage I, stage II, stage III, stage IV) (Fig. 3a–c). The expression level of B7-1 was significantly lower while the expression level for B7-H3 was higher in patients with tumor after treatment than in patients who are tumor free in LUAD (Fig. 3a). Moreover, the expression of B7-1/H1/H2/H4/H5 was significantly lower in male than in female in LUAD. Meanwhile, the expression of B7-1/2/H5 was significantly lower in male than in female in LUSC (Fig. 3b). For pathological stage, the expression level of B7-H3 exhibit significant difference in LUAD increasing from stage I to stage III but decreasing in stage IV. Compared with stage I, the expression of B7-H1 was decreased and B7-H4 was increased in stage IV in LUSC (Fig. 3c).
The association ofB7 family expression with clinicopathological parametersand patient survival. The relationship between the expression of B7 family members and (a) cancer status (*P < 0.05; **P < 0.01 and ***P < 0.001), (b) gender and (c) pathologic stages (Stage I, Stage II, Stage III, Stage IV). (d) Kaplan–Meier survival curves for different subtypes of NSCLC patients stratified according to high vs low expression of B7 family.
Subsequently, we also explored the relationship between expression of each B7 family member with overall survival (OS) (Fig. 3d). According to Kaplan–Meier analysis, the expression of B7-1/2/H3 was associated with patient survival in LUAD and the expression of B7-H5 was associated with patient survival in LUSC. In LUAD, higher expression of B7-1 and B7-2 was associated with prolonged OS while elevated expression of B7-H3 was associated with worse OS. In LUSC, higher expression of B7-H5 predicted shortened OS. Consistent with our finding, high expression level of B7-H3 and B7-H5 has been reported to be associated with poor OS in NSCLC23,24.
Function and pathway analysis for differentially expressed proteins
Changes in protein expression caused by B7 family gene alterations were extracted from the Reverse Phase Protein Arrays (RPPA) database and analyzed in cBioportal. Significantly changed proteins in LUAD and LUSC were screened out in RPPA, respectively (Fig. 4a). The intersection of the top10 most significant signaling pathways enriched by different B7 family members was shown in Fig. 4b. The ErbB signaling pathway and EGFR tyrosine kinase inhibitor resistance were the most significant pathway influenced by B7 member alteration in both LUAD and LUSC (Fig. 4b). PI3K-Akt signaling pathway and Non-small cell lung cancer were also among the top pathways that the differentially expressed proteins were involved in for both NSCLC subtypes. Interestingly, these significant pathways all collectively presented in the ErbB signaling pathway (Supplementary Fig. 1)25,26,27. At the same time, the three major GO(Gene Ontology) terms for the differentially expressed proteins were also analyzed (Fig. 4c). The results for LUAD and LUSC were very similar, with protein binding being the major molecular function for the differentially expressed proteins. Signal transduction was the most important biological process and the most significant cellular component was cytosol.
GO and KEGG enrichment analysis of B7 family. (a) Enrichmentof differentially expressed proteins of B7 family in NSCLC. − log10 (P value) = 1.30 is considered to be cut-off value for difference. The distance of expression level change was based on logarithmic ratio (mean in altered/mean in unaltered). (b) Intersection of top10 important KEGG signaling pathways for each B7 family member. (c) The influence of B7 family on downstream processes were analyzed by GO enrichment analysis via DAVID.
MAPK1/3 and MAP2K1 are important B7 family member related genes in NSCLC
For a better understanding of the interactions amongst these differential proteins in the EGFR tyrosine kinase inhibitor resistance and ErbB signaling pathway and to identify the most important proteins, the functional network diagram of the top 10 important proteins were identified using Cytoscape(Fig. 5a, b). These crucial proteins displayed complex interactions in both PI3K-Akt signaling pathway and MAPK signaling pathway. At the same time, GOSemSim and org.hs.eg.db R package were used to calculate the similarities of the differentially expressed proteins in the EGFR tyrosine kinase inhibitor resistance and ErbB signaling pathway by considering the GO topology (Fig. 5c, d). Moreover, MAPK1/3 and MAP2K1 exhibited high functional similarities among these differentially expressed proteins. The expression levels of B7 family and these differentially expressed protein-coding genes were utilized for calculating the correlation (Fig. 5e, f). Overall, the cascade formed by the expression of genes related to the PI3K-Akt signaling pathway and MAPK signaling pathway also exhibited extremely high internal correlation. MAPK1/3 and MAP2K1 were found to be important differential proteins in both LUAD and LUSC and exhibited good correlation with B7 members. MAPK1/3 and MAP2K1 were positively and significantly correlated with B7-H2/H3/H4/H5 in LUAD and also positively and significantly correlated with B7-H3/H6 in LUSC. Intriguingly, MAPK1/3 and MAP2K1 were critical components of ErbB/MAPK signaling pathway as well (Fig S1a). All of this evidence suggests that the actions of the ErbB/MAPK signaling pathway and ErbB/PI3K-Akt signaling pathway would be affected by the regulation of the B7 family.
Identification of pivotal B7 family related genes in NSCLC. (a, b) Cytoscape identified top10 important differentially expressed proteins regulated by B7 family in both LUAD and LUSC.The intensity of color showed the importance of genes in the networks. (c, d) Similarity of differentially expressed proteins based on Gene ontology in different GO terms inLUAD and LUSC. (e, f) Correlation analysis of differentially expressed protein-coding genes enriched in the PI3K-Akt signaling pathway and MAPK signaling pathway in LUAD and LUSC. Here, Pearson correlation coefficient was used to quantify the correlation.
Discussion
B7 family members act as the secondary co-stimulatory or co-inhibitory signals regulating T cell activation and play an important role in the immune response to NK cells15. In terms of the developing two-signal theory of T cells, the binding action of T cell receptor (TCR) alone does not activate T cells but signals from co-stimulating and co-suppressing molecules can regulate TCR signals14. For example, signals from the B7 family and its ligand CD28 family can bind to TCR to affect the strength and duration of the immune response, which not only plays adramatic role in the anti-tumor process but can also maintain its own tolerance and thus inhibits the autoimmunity13,14,28. Expressions of some B7 family members are associated with the development of a number of diseases, such as extramammary Paget disease (EMPD), diffuse large B-cell lymphoma (DLBCL), renal cancer and advanced gastric cancer, suggesting its importance in tumorigenicity and immune escape15,29,30. Due to the unclear expression pattern, regulation mechanism and downstream signal transduction pathway of B7 family in NSCLC, we herein provide a comprehensive study of B7 family using a variety of bioinformatics analysis.
In the present study, we first examined the expression profile of B7 family in NSCLC (Fig. 1a, b). There were consistently increased levels of B7-H3/H4 and decreased B7-1/2/DC/H1/H2/H5/H6 in the two NSCLC subtypes. B7-1 (CD80) and B7-2 (CD86) are co-stimulatory molecules which triggers T cell activation. Low expression of B7-1 and B7-2 has been found in different tumors and positivity of them are usually associated with better OS31,32,33, which is also found in our analysis. B7-H2 is a co-stimulatory member of the B7 family and interaction of B7-H2 and its receptor ICOS promotes T cell responses. However, the role of B7-H2 in cancer is complicated. On one hand, B7-H2 expressing myeloma cells induced CD4+ T cells to proliferate and produce soluble factors which stimulate myeloma cell proliferation34. On the other hand, ICOS/B7-H2 signal enhances secondary responses by CD8(+) T cells and improves effectiveness of cancer therapy35,36. Nonetheless, selective down-regulation of B7-H2 on tumor cells is deemed as a means of tumor immune escape37,38. B7-H3 and B7-H4 has been widely reported to be over-expressed in different cancers including lung cancer15,23,39,40 and their high expression was correlated with poor overall survival23. NSCLC tissues expressing B7-H3 or B7-H4 had fewer T infiltrating lymphoid cells (TILs) and significantly more common lymph node metastasis41. B7-H5 is a ligand for CD28H and acts as a co-inhibitory molecule. The expression of B7-H5 is predominantly found in hematopoietic tissues, Treg cells and myeloid cells15,42. B7-H5 is also widely expressed in tumor cells and high expression of it is related to a worse prognosis in various cancers including lung cancer24. It is well known that EGFR is notorious for being widely expressed and mutated in NSCLC43. It participates in immune evasion, most likely by regulating the expression of B7-H544. B7-H6 is a ligand of NK cell activating receptor NKp30. B7-H6 can be induced by inflammatory stress in healthy cells, and is also expressed on various primary human tumors15,45. Although abnormal over-expression of B7-H6 was linked with poor prognosis in various types of cancers46,47 its expression in NSCLC is limited48.
Promoter DNA methylation, as an important epigenetic modification, has a widespread association with gene expression49,50. Therefore, we further investigated whether promoter DNA methylation were involved in B7 member dysregulation. As shown in Fig. 1c, the expression of most B7 members were significantly and negatively correlated with promoter DNA methylation in LUAD while only B7-H1 and B7-H4 were significantly associated with DNA methylation in LUSC. On the other hand, the expression of almost all B7 members was significantly associated with copy number alterations (Fig. 1d). These results suggested that both DNA methylation and copy number alterations might be involved in the altered expression of B7 members in NSCLC. Similar with our results, promoter methylation has also been reported to show regulatory effect on the expression of B7-1/2/H3/H4/H5/H6/H7 in previous studies51,52,53. Since the expression of B7 members was significantly related to copy number alterations, we then further determined the mutation frequency and mutation rate of B7 family members in NSCLC (Fig. 2a, b), including mutation, amplification and deep deletion. The overall mutation frequency and mutation rate of B7 family in LUSC were higher than that of LUAD. The mutation rates were relatively higher for B7-1and B7-2 in LUSC and also for B7-DC, B7-H1 and B7-H4 in both subtypes. Amplification accounted for the majority of mutation in B7 members with increased expression, such as B7-1, B7-2, B7-H3, B7-H5, B7-H6 in both NSCLC subtypes and B7-H2 and B7-H7 in LUSC. Deep deletion was frequently found in B7-DC, B7-H1 and B7-H4 in LUAD and LUSC. Consistent with our results, gene amplification of B7-H1 (PD-L1) has been demonstrated by several studies54,55,56 and was associated with response to nivolumab monotherapy among patients with NSCLC56. Shallow or deep deletion of B7-H1 was reported in head and neck cancer57. Deep deletion was also a major mutation type for B7-DC and B7-H1 in gastric cancer53 and breast cancer47 and for B7-H4 in signet ring cell carcinoma of the stomach53.
To find the potential targets and downstream pathways of B7 family, differentially expressed proteins influenced by B7 family were identified and subjected to KEGG and GO enrichment analysis (Fig. 4a–c). Intriguingly, these differential protein-coding genes were commonly enriched within ErbB signaling pathway and three GO processes mainly involving protein binding, cytosol and signal transduction in both NSCLC subtypes. ErbB signaling pathway is frequently overly activated in malignancies of epithelial origin and signaling from ErbB receptors engaged in every cellular process including proliferation, survival, migration and differentiation and cell and tissue morphogenesis58. Epithelial growth factor receptor (EGFR also ERBB1or HER1) has been the most well studied of the four members of the ErbB family. Over 60% of NSCLC patients express EGFR and tyrosine kinase inhibitors (TKIs) targeting the kinase domain of EGFR are clinically effective in NSCLC patients harboring activating mutations in the tyrosine kinase domain of the EGFR gene43. The role of EGFR in regulating PD-L1 expression in NSCLC has been reported by several studies59,60,61. One study found that activation of EGFR and mutations of 19 deletion or L858R induced PD-L1 expression via the p-ERK1/2/p-c-Jun signaling pathway61. The other study revealed that EGFR up-regulated PD-L1 expression through the IL-6/JAK/STAT3 signaling pathway62. Both studies found that treatment with EGFR TKIs reduced the expression of PD-L1. However, the clinical efficacy of anti-PD-1/PD-L1 immune checkpoint inhibitors (ICIs) in advanced EGFR mutant NSCLC or combination therapy of EGFR-TKIs and anti-PD-1/PD-L1 ICIs is still not clear yet63. Research evidence and clinical trials showed that combination of anti-PD-1/PD-L1 ICIs and EGFR TKIs did not exhibit synergistic anticancer effect in NSCLC61,63. It is suggested that anti-PD-1/PD-L1 ICIs in combination with chemotherapy or other targeted therapy might be more effective in treating advanced NSCLC63. In addition to EGFR, research evidence showed that ERBB2 and ERBB3 mutation could also upregulate PD-L1 expression and suppress normal T-cell-mediated cytotoxicity through activation of the PI3K/Akt signaling pathway64. Together, our findings stress a close relation between ErbB signaling and B7 family expression, which is partially supported by the literature and awaits further investigation.
At the same time, our results demonstrated that MAP2K1 (MEK), MAPK1 (ERK, also ERK2) and MAPK3 (ERK1) were important B7 family related genes in both NSCLC subtypes and were positively correlated with several B7 family members (Fig. 5). The activation of MEK/ERK pathway has been reported for different B7 members. According to the literature, B7-H4 contribute to cancer progression by activation of the ERK 1/2 signaling65,66. B6-H6 could also promote non-hodgkin lymphoma via Ras/MEK/ERK pathway67. B7-H3 regulates breast cancer stemness by activating MEK/MAPK signaling68. In colorectal cancer, PD-L1 upregulates HMGA1 to activate PI3K/Akt and MEK/ERK pathways to promote cancer stem cell expansion69. On the other hand, the ERK-MAPK pathway has been shown to regulate PD-L1 expression in different cancer types70,71,72. ERK and JNK pathways are also involved in IFN-γ-induced B7-DC expression on tumor cells73. Interestingly, out findings revealed a strong positive correlation between MEK/ERK and B7-H3 and B7-H6 in NSCLC which has been rarely studied. One study showed that combination therapy of MEK inhibitor with B7-H3-redirected bispecific antibody significantly suppressed in vivo tumor growth and increased T cell infiltration74. Synergistic antitumor effect has also been observed for MEK inhibitors combing with anti-PD-1 or anti-PD-L1 antibodies in different ex-vivo and in vivo models in various cancers75,76,77,78. The use of compounds targeting Ras-Raf-MEK-ERK signaling pathway, such as RAF or MEK inhibitors, has led to substantial improvement in clinical outcome in various types of tumor75. However, resistance to these agents is usually found. Several orally effective, potent, and specific inhibitors of ERK1/2 are in early clinical trials79. These results underline the potential of combination treatment combining MEK or ERK inhibitors with PD-1/PD-L1/B7-H3/B7-H6 blockade.
To date, the epidermal growth factor receptor (EGFR/ERBB1) distinctive features of the ERBB family provides a foundation for the design of therapies targeting NSCLC58,80. Tyrosine kinase inhibitors (TKIs) treatment has been constantly updated and combined with other treatment options to improve prognosis of NSCLC80,81. The Shc-activated or Grb2-activated mitogen-activated protein kinase (MAPK) pathway is a common downstream signaling of ErbB receptors (Fig. S1c). MAP2K1 (MEK), MAPK1 (ERK) and MAPK3 (ERK1)play a pivotal role in regulating cell proliferation, transcriptional regulation, differentiation and survival as members of the MAPK signaling pathway82. Collectively, our study supports a close link between B7 family and the ErbB/MAPK signaling pathway in NSCLC. More in-depth exploration of TKIs or MEK/ERK inhibitors combining with B7 checkpoint inhibitors will be help for designing rational immunotherapy for NSCLC.
Methods
Data analysis
Data were extracted from The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov/) and Genotype-Tissue Expression (GTEx) (https://www.gtexportal.org/home/), expression levels of B7 family members were log2-transformed and matched clinical data in NSCLC.GISTIC 2.0 database and reversedphase protein array(RPPA) from cBioportal website (https://www.cbioportal.org/) were used for data visualization, such as somatic mutations, copy number variations, DNA methylation and screening of differentially expressed proteins83. The relationship between mRNA expression levels of B7 family members and DNA methylation or copy number changes was analyzed using cBioportal database.
Differential analysis
Differential analysis of patients with LUAD and LUSC according to whether each B7 family member is mutated on the cBioportal website. P value < 0.05 is identified as differentially expressed proteins. All information of differential analysis corresponding to each B7 family member is deposited in Supplementary table 1 and 2.
Protein and functional network analysis
The PPI network is constructed using STRING and Cytoscape software. The STRING online database (https://string-db.org/) has powerful visualization and customization capabilities to annotate protein–protein interactions (PPI) network84. CytoHubba plugin of Cytoscape provides 11 topological analysis methods to identify important nodes in biological networks85.
GO and KEGG enrichment analysis
The screened differentially expressed proteins were subjected to Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis in the DAVID function annotation tool (https://david.ncifcrf.gov). DAVID is a database of biological information that integrates biological data and analysis tools to provide systematic and comprehensive biological functional annotation information for large-scale gene or protein lists (hundreds of genes or protein ID lists)86,87. The abundance of biological processes (BP), molecular functions(MF), and cellular components(CC) was analyzed, and the influenced metabolic processes were obtained by consideration of gene counts and P value.
R project analysis
Kaplan–Meier analysis was performed using survminer R package. Comparison of functional similarity of BP, MF and CC between proteins was constructed using GOSemSim and org.hs.eg.db packages. The R package of GOSemSim depended on GO annotation information to compute the semantic similarity among GO terms, gene products, and gene clusters88. From this analysis, major genes contributed to GO analysis can be found. Corrplot package was used to characterize the extent of co-expression between differentially expressed genes by computing gene expression. The ggpubr package was utilized to display mRNA expression levels in normal and tumor tissues.
Statistical analysis
GraphPad Prism 7 (GraphPad Software, La Jolla, CA) was used for statistical analysis. Student's t-test was utilized to compare differences between two groups. (*P < 0.05; **P < 0.01 and ***P < 0.001) One-way ANOVA was utilized to compare multiple groups. Spearman's and Pearson's correlation analysis was utilized to calculate the correlation between the two groups. Kaplan–Meier analysis was used to analyze the overall survival rate based on TCGA data, and the P value was calculated by log-rank tests. P < 0.05 was considered as statistically significant.
Data availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
References
Schabath, M. B. & Cote, M. L. Cancer progress and priorities: Lung cancer. Cancer Epidemiol. Biomark. Prev. 28, 1563–1579. https://doi.org/10.1158/1055-9965.EPI-19-0221 (2019).
Nagasaka, M. & Gadgeel, S. M. Role of chemotherapy and targeted therapy in early-stage non-small cell lung cancer. Expert Rev. Anticancer Ther. 18, 63–70. https://doi.org/10.1080/14737140.2018.1409624 (2018).
Brown, S., Banfill, K., Aznar, M. C., Whitehurst, P. & Faivre Finn, C. The evolving role of radiotherapy in non-small cell lung cancer. Br. J. Radiol. 92, 20190524. https://doi.org/10.1259/bjr.20190524 (2019).
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Tye, L. S., Scott, T., Haszard, J. J. & Peddie, M. C. Physical activity, sedentary behaviour and sleep, and their association with BMI in a sample of adolescent females in New Zealand. Int. J. Environ. Res. Public Health https://doi.org/10.3390/ijerph17176346 (2020).
Ruiz-Cordero, R. & Devine, W. P. Targeted therapy and checkpoint immunotherapy in lung cancer. Surg. Pathol. Clin. 13, 17–33. https://doi.org/10.1016/j.path.2019.11.002 (2020).
Herbst, R. S., Morgensztern, D. & Boshoff, C. The biology and management of non-small cell lung cancer. Nature 553, 446–454. https://doi.org/10.1038/nature25183 (2018).
Marwitz, S. et al. The multi-modal effect of the anti-fibrotic drug pirfenidone on NSCLC. Front. Oncol. 9, 1550. https://doi.org/10.3389/fonc.2019.01550 (2019).
Duma, N., Santana-Davila, R. & Molina, J. R. Non-small cell lung cancer: Epidemiology, screening, diagnosis, and treatment. Mayo Clin. Proc. 94, 1623–1640. https://doi.org/10.1016/j.mayocp.2019.01.013 (2019).
Liu, X., Liu, J., Zhang, X., Tong, Y. & Gan, X. MiR-520b promotes the progression of non-small cell lung cancer through activating Hedgehog pathway. J. Cell Mol. Med. 23, 205–215. https://doi.org/10.1111/jcmm.13909 (2019).
Ashour Badawy, A. et al. Site of metastases as prognostic factors in unselected population of stage IV non-small cell lung cancer. Asian Pac. J. Cancer Prev. 19, 1907–1910. https://doi.org/10.22034/APJCP.2018.19.7.1907 (2018).
Yuan, L., Ye, J. & Fan, D. The B7–H4 gene induces immune escape partly via upregulating the PD-1/Stat3 pathway in non-small cell lung cancer. Hum. Immunol. 81, 254–261. https://doi.org/10.1016/j.humimm.2020.02.004 (2020).
Azuma, M. Co-signal molecules in T-cell activation: Historical overview and perspective. Adv. Exp. Med. Biol. 1189, 3–23. https://doi.org/10.1007/978-981-32-9717-3_1 (2019).
Yang, X., Lin, G., Han, Z. & Chai, J. Structural biology of NOD-like receptors. Adv. Exp. Med. Biol. 1172, 119–141. https://doi.org/10.1007/978-981-13-9367-9_6 (2019).
Ni, L. & Dong, C. New B7 family checkpoints in human cancers. Mol. Cancer Ther. 16, 1203–1211. https://doi.org/10.1158/1535-7163.MCT-16-0761 (2017).
Zong, L. et al. PD-L1, B7–H3 and VISTA are highly expressed in gestational trophoblastic neoplasia. Histopathology 75, 421–430. https://doi.org/10.1111/his.13882 (2019).
Zong, Z. et al. M1 macrophages induce PD-L1 expression in hepatocellular carcinoma cells through IL-1beta signaling. Front. Immunol. 10, 1643. https://doi.org/10.3389/fimmu.2019.01643 (2019).
He, L. & Li, Z. B7–H3 and its role in bone cancers. Pathol. Res. Pract. 215, 152420. https://doi.org/10.1016/j.prp.2019.04.012 (2019).
Sharma, P., Wagner, K., Wolchok, J. D. & Allison, J. P. Novel cancer immunotherapy agents with survival benefit: Recent successes and next steps. Nat. Rev. Cancer 11, 805–812. https://doi.org/10.1038/nrc3153 (2011).
Shima, T. et al. Infiltration of tumor-associated macrophages is involved in tumor programmed death-ligand 1 expression in early lung adenocarcinoma. Cancer Sci. 111, 727–738. https://doi.org/10.1111/cas.14272 (2020).
Song, F. et al. Identification of key microRNAs and hub genes in non-small-cell lung cancer using integrative bioinformatics and functional analyses. J. Cell Biochem. 121, 2690–2703. https://doi.org/10.1002/jcb.29489 (2020).
Naylor, E. C., Desani, J. K. & Chung, P. K. Targeted therapy and immunotherapy for lung cancer. Surg. Oncol. Clin. N. Am. 25, 601–609. https://doi.org/10.1016/j.soc.2016.02.011 (2016).
Altan, M. et al. B7–H3 expression in NSCLC and its association with B7–H4, PD-L1 and tumor-infiltrating lymphocytes. Clin. Cancer Res. 23, 5202–5209. https://doi.org/10.1158/1078-0432.CCR-16-3107 (2017).
Zhong, C. et al. Phenotypical and potential functional characteristics of different immune cells expressing CD28H/B7-H5 and their relationship with cancer prognosis. Clin. Exp. Immunol. 200, 12–21. https://doi.org/10.1111/cei.13413 (2020).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Ishiguro-Watanabe, M. & Tanabe, M. KEGG: Integrating viruses and cellular organisms. Nucleic Acids Res. 49, D545–D551. https://doi.org/10.1093/nar/gkaa970 (2021).
Zheng, S. et al. A B7-CD28 family based signature demonstrates significantly different prognoses and tumor immune landscapes in lung adenocarcinoma. Int. J. Cancer 143, 2592–2601. https://doi.org/10.1002/ijc.31764 (2018).
Pourmaleki, M. et al. Extramammary Paget disease shows differential expression of B7 family members B7–H3, B7–H4, PD-L1, PD-L2 and cancer/testis antigens NY-ESO-1 and MAGE-A. Oncotarget 10, 6152–6167. https://doi.org/10.18632/oncotarget.27247 (2019).
Wang, G. et al. B7-CD28 gene family expression is associated with prognostic and immunological characteristics of diffuse large B-cell lymphoma. Aging (Albany NY) 11, 3939–3957. https://doi.org/10.18632/aging.102025 (2019).
Feng, X. Y. et al. Low expression of CD80 predicts for poor prognosis in patients with gastric adenocarcinoma. Future Oncol. 15, 473–483. https://doi.org/10.2217/fon-2018-0420 (2019).
Tatsumi, T. et al. Expression of costimulatory molecules B7–1 (CD80) and B7–2 (CD86) on human hepatocellular carcinoma. Hepatology 25, 1108–1114. https://doi.org/10.1002/hep.510250511 (1997).
Chang, C. S., Chang, J. H., Hsu, N. C., Lin, H. Y. & Chung, C. Y. Expression of CD80 and CD86 costimulatory molecules are potential markers for better survival in nasopharyngeal carcinoma. BMC Cancer 7, 88. https://doi.org/10.1186/1471-2407-7-88 (2007).
Yamashita, T. et al. Functional B7.2 and B7–H2 molecules on myeloma cells are associated with a growth advantage. Clin. Cancer Res. 15, 770–777. https://doi.org/10.1158/1078-0432.CCR-08-0501 (2009).
Wallin, J. J., Liang, L., Bakardjiev, A. & Sha, W. C. Enhancement of CD8+ T cell responses by ICOS/B7h costimulation. J. Immunol. 167, 132–139. https://doi.org/10.4049/jimmunol.167.1.132 (2001).
Marinelli, O. et al. ICOS-L as a potential therapeutic target for cancer immunotherapy. Curr. Protein Pept. Sci. 19, 1107–1113. https://doi.org/10.2174/1389203719666180608093913 (2018).
Richter, G. & Burdach, S. ICOS: A new costimulatory ligand/receptor pair and its role in T-cell activion. Onkologie 27, 91–95. https://doi.org/10.1159/000075612 (2004).
Flies, D. B. & Chen, L. The new B7s: Playing a pivotal role in tumor immunity. J. Immunother. 30, 251–260. https://doi.org/10.1097/CJI.0b013e31802e085a (2007).
Zang, X. et al. B7–H3 and B7x are highly expressed in human prostate cancer and associated with disease spread and poor outcome. Proc. Natl. Acad. Sci. USA 104, 19458–19463. https://doi.org/10.1073/pnas.0709802104 (2007).
Tringler, B. et al. B7–h4 is highly expressed in ductal and lobular breast cancer. Clin. Cancer Res. 11, 1842–1848. https://doi.org/10.1158/1078-0432.CCR-04-1658 (2005).
Sun, Y. et al. B7–H3 and B7–H4 expression in non-small-cell lung cancer. Lung Cancer 53, 143–151. https://doi.org/10.1016/j.lungcan.2006.05.012 (2006).
Andrews, L. P., Yano, H. & Vignali, D. A. A. Inhibitory receptors and ligands beyond PD-1, PD-L1 and CTLA-4: Breakthroughs or backups. Nat. Immunol. 20, 1425–1434. https://doi.org/10.1038/s41590-019-0512-0 (2019).
da Cunha Santos, G., Shepherd, F. A. & Tsao, M. S. EGFR mutations and lung cancer. Annu. Rev. Pathol. 6, 49–69. https://doi.org/10.1146/annurev-pathol-011110-130206 (2011).
Dong, Z., Zhang, L., Xu, W. & Zhang, G. EGFR may participate in immune evasion through regulation of B7H5 expression in nonsmall cell lung carcinoma. Mol. Med. Rep. 18, 3769–3779. https://doi.org/10.3892/mmr.2018.9361 (2018).
Che, F. et al. B7–H6 expression is induced by lipopolysaccharide and facilitates cancer invasion and metastasis in human gliomas. Int. Immunopharmacol. 59, 318–327. https://doi.org/10.1016/j.intimp.2018.03.020 (2018).
Hu, Y. et al. Immunological role and underlying mechanisms of B7–H6 in tumorigenesis. Clin. Chim. Acta 502, 191–198. https://doi.org/10.1016/j.cca.2019.12.030 (2020).
Xu, Z. et al. Comprehensive molecular profiling of the B7 family of immune-regulatory ligands in breast cancer. Oncoimmunology 5, e1207841. https://doi.org/10.1080/2162402X.2016.1207841 (2016).
Zhang, X., Zhang, G., Qin, Y., Bai, R. & Huang, J. B7–H6 expression in non-small cell lung cancers. Int. J. Clin. Exp. Pathol. 7, 6936–6942 (2014).
Jones, M. J., Goodman, S. J. & Kobor, M. S. DNA methylation and healthy human aging. Aging Cell 14, 924–932. https://doi.org/10.1111/acel.12349 (2015).
Yonesaka, K. et al. B7–H3 negatively modulates CTL-mediated cancer immunity. Clin. Cancer Res. 24, 2653–2664. https://doi.org/10.1158/1078-0432.CCR-17-2852 (2018).
Wang, Z. et al. Genetic and clinical characterization of B7–H3 (CD276) expression and epigenetic regulation in diffuse brain glioma. Cancer Sci. 109, 2697–2705. https://doi.org/10.1111/cas.13744 (2018).
de Vos, L. et al. The landscape of CD28, CD80, CD86, CTLA4, and ICOS DNA methylation in head and neck squamous cell carcinomas. Epigenetics 15, 1195–1212. https://doi.org/10.1080/15592294.2020.1754675 (2020).
Li, D. et al. Comprehensive understanding of B7 family in gastric cancer: Expression profile, association with clinicopathological parameters and downstream targets. Int. J. Biol. Sci. 16, 568–582. https://doi.org/10.7150/ijbs.39769 (2020).
Straub, M. et al. CD274/PD-L1 gene amplification and PD-L1 protein expression are common events in squamous cell carcinoma of the oral cavity. Oncotarget 7, 12024–12034. https://doi.org/10.18632/oncotarget.7593 (2016).
Saito, R. et al. Overexpression and gene amplification of PD-L1 in cancer cells and PD-L1(+) immune cells in Epstein–Barr virus-associated gastric cancer: The prognostic implications. Mod. Pathol. 30, 427–439. https://doi.org/10.1038/modpathol.2016.202 (2017).
Inoue, Y. et al. Evaluation of programmed death ligand 1 (PD-L1) gene amplification and response to nivolumab monotherapy in non-small cell lung cancer. JAMA Netw. Open 3, e2011818. https://doi.org/10.1001/jamanetworkopen.2020.11818 (2020).
Chen, Y. P. et al. The immune molecular landscape of the B7 and TNFR immunoregulatory ligand-receptor families in head and neck cancer: A comprehensive overview and the immunotherapeutic implications. Oncoimmunology 6, e1288329. https://doi.org/10.1080/2162402X.2017.1288329 (2017).
Aran, V. & Omerovic, J. Current approaches in NSCLC targeting K-RAS and EGFR. Int. J. Mol. Sci. https://doi.org/10.3390/ijms20225701 (2019).
Akbay, E. A. et al. Activation of the PD-1 pathway contributes to immune escape in EGFR-driven lung tumors. Cancer Discov. 3, 1355–1363. https://doi.org/10.1158/2159-8290.CD-13-0310 (2013).
Azuma, K. et al. Association of PD-L1 overexpression with activating EGFR mutations in surgically resected nonsmall-cell lung cancer. Ann. Oncol. 25, 1935–1940. https://doi.org/10.1093/annonc/mdu242 (2014).
Chen, N. et al. Upregulation of PD-L1 by EGFR activation mediates the immune escape in EGFR-driven NSCLC: Implication for optional immune targeted therapy for NSCLC patients with EGFR mutation. J. Thorac. Oncol. 10, 910–923. https://doi.org/10.1097/JTO.0000000000000500 (2015).
Zhang, N. et al. The EGFR pathway is involved in the regulation of PD-L1 expression via the IL-6/JAK/STAT3 signaling pathway in EGFR-mutated non-small cell lung cancer. Int. J. Oncol. 49, 1360–1368. https://doi.org/10.3892/ijo.2016.3632 (2016).
Hsu, P. C., Jablons, D. M., Yang, C. T. & You, L. Epidermal growth factor receptor (EGFR) pathway, yes-associated protein (YAP) and the regulation of programmed death-ligand 1 (PD-L1) in non-small cell lung cancer (NSCLC). Int. J. Mol. Sci. https://doi.org/10.3390/ijms20153821 (2019).
Li, M. et al. Genomic ERBB2/ERBB3 mutations promote PD-L1-mediated immune escape in gallbladder cancer: A whole-exome sequencing analysis. Gut 68, 1024–1033. https://doi.org/10.1136/gutjnl-2018-316039 (2019).
Xie, N. et al. Upregulation of B7–H4 promotes tumor progression of intrahepatic cholangiocarcinoma. Cell Death Dis. 8, 3205. https://doi.org/10.1038/s41419-017-0015-6 (2017).
Jiang, Y. et al. B7–H4 is highly expressed in aggressive Epstein–Barr virus positive diffuse large B-cell lymphoma and inhibits apoptosis through upregulating Erk1/2 and Akt signalling pathways. Infect. Agent Cancer 14, 20. https://doi.org/10.1186/s13027-019-0234-9 (2019).
Yang, S. et al. B7–H6 promotes cell proliferation, migration and invasion of non-hodgkin lymphoma via Ras/MEK/ERK pathway based on quantitative phosphoproteomics data. Onco Targets Ther. 13, 5795–5805. https://doi.org/10.2147/OTT.S257512 (2020).
Liu, Z. et al. Immunoregulatory protein B7–H3 regulates cancer stem cell enrichment and drug resistance through MVP-mediated MEK activation. Oncogene 38, 88–102. https://doi.org/10.1038/s41388-018-0407-9 (2019).
Wei, F. et al. PD-L1 promotes colorectal cancer stem cell expansion by activating HMGA1-dependent signaling pathways. Cancer Lett. 450, 1–13. https://doi.org/10.1016/j.canlet.2019.02.022 (2019).
Yamamoto, R. et al. B7–H1 expression is regulated by MEK/ERK signaling pathway in anaplastic large cell lymphoma and Hodgkin lymphoma. Cancer Sci. 100, 2093–2100. https://doi.org/10.1111/j.1349-7006.2009.01302.x (2009).
Sumimoto, H., Takano, A., Teramoto, K. & Daigo, Y. RAS-mitogen-activated protein kinase signal is required for enhanced PD-L1 expression in human lung cancers. PLoS ONE 11, e0166626. https://doi.org/10.1371/journal.pone.0166626 (2016).
Su, L. et al. EGFR-ERK pathway regulates CSN6 to contribute to PD-L1 expression in glioblastoma. Mol. Carcinog. 59, 520–532. https://doi.org/10.1002/mc.23176 (2020).
Deng, J. et al. Involvement of ERK and JNK pathways in IFN-gamma-induced B7-DC expression on tumor cells. J. Cancer Res. Clin. Oncol. 137, 243–250. https://doi.org/10.1007/s00432-010-0876-x (2011).
Li, H. et al. MEK inhibitor augments antitumor activity of B7–H3-redirected bispecific antibody. Front. Oncol. 10, 1527. https://doi.org/10.3389/fonc.2020.01527 (2020).
Samatar, A. A. & Poulikakos, P. I. Targeting RAS-ERK signalling in cancer: Promises and challenges. Nat. Rev. Drug Discov. 13, 928–942. https://doi.org/10.1038/nrd4281 (2014).
Lee, J. W. et al. The combination of MEK inhibitor with immunomodulatory antibodies targeting programmed death 1 and programmed death ligand 1 results in prolonged survival in Kras/p53-driven lung cancer. J. Thorac. Oncol. 14, 1046–1060. https://doi.org/10.1016/j.jtho.2019.02.004 (2019).
Liu, L. et al. The BRAF and MEK inhibitors dabrafenib and trametinib: Effects on immune function and in combination with immunomodulatory antibodies targeting PD-1, PD-L1, and CTLA-4. Clin. Cancer Res. 21, 1639–1651. https://doi.org/10.1158/1078-0432.CCR-14-2339 (2015).
Napolitano, S. et al. Triple blockade of EGFR, MEK and PD-L1 has antitumor activity in colorectal cancer models with constitutive activation of MAPK signaling and PD-L1 overexpression. J. Exp. Clin. Cancer Res. 38, 492. https://doi.org/10.1186/s13046-019-1497-0 (2019).
Roskoski, R. Jr. Targeting ERK1/2 protein-serine/threonine kinases in human cancers. Pharmacol. Res. 142, 151–168. https://doi.org/10.1016/j.phrs.2019.01.039 (2019).
Remon, J., Steuer, C. E., Ramalingam, S. S. & Felip, E. Osimertinib and other third-generation EGFR TKI in EGFR-mutant NSCLC patients. Ann. Oncol. 29, i20–i27. https://doi.org/10.1093/annonc/mdx704 (2018).
Rebuzzi, S. E. et al. Combination of EGFR-TKIs and chemotherapy in advanced EGFR mutated NSCLC: Review of the literature and future perspectives. Crit. Rev. Oncol. Hematol. 146, 102820. https://doi.org/10.1016/j.critrevonc.2019.102820 (2020).
Scheffler, M. et al. Co-occurrence of targetable mutations in Non-small cell lung cancer (NSCLC) patients harboring MAP2K1 mutations. Lung Cancer 144, 40–48. https://doi.org/10.1016/j.lungcan.2020.04.020 (2020).
Gao, J. et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal 6, pl1. https://doi.org/10.1126/scisignal.2004088 (2013).
Szklarczyk, D. et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 47, D607–D613. https://doi.org/10.1093/nar/gky1131 (2019).
Chin, C. H. et al. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 8(Suppl 4), S11. https://doi.org/10.1186/1752-0509-8-S4-S11 (2014).
Huang, D. W. et al. DAVID bioinformatics resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 35, W169-175. https://doi.org/10.1093/nar/gkm415 (2007).
da Huang, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57. https://doi.org/10.1038/nprot.2008.211 (2009).
Yu, G. et al. GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26, 976–978. https://doi.org/10.1093/bioinformatics/btq064 (2010).
Funding
This work was supported by National Natural Science Foundation of China (No. 81972643), Sichuan Science and Technology Project (2021YJ0201) and Luxian People's Government and Southwest Medical University Scientific and Technological Achievements Transfer and Transformation Strategic Cooperation Project (2019LXXNYKD-07) and China Center for Evidence Based Traditional Chinese Medicine (2019XZZX-ZL005).
Author information
Authors and Affiliations
Contributions
M.X. and C.P. clutched the article and designed the research idea. Y.Z., X.W., M.L., F.W. and Y.C. collected and organized the study data, and preprocessed the data in statistical software. F.D. and S.X. visualized the results and embellished and typeset the image in the software. Q.W., Z.Y., Z.X. and J.S. jointly revised the article. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xiao, M., Pang, C., Xiang, S. et al. Comprehensive characterization of B7 family members in NSCLC and identification of its regulatory network. Sci Rep 13, 4311 (2023). https://doi.org/10.1038/s41598-022-26776-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-26776-w
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.