Abstract
The gene encoding beta2-adrenergic receptor (β2-AR), adrenoceptor beta 2 (ADRB2), has been reported to closely associated with various cancers. However, its role in lung adenocarcinoma (LUAD) remains controversial. This research shed light on the prognostic value of ADRB2 in LUAD and further explored its association with immune cell infiltration. ADRB2 was significantly decreased in LUAD. ADRB2 expression in LUAD was significantly correlated with gender, smoking status, T classification, and pathologic stage. Patients in the low ADRB2 expression group presented with significantly poorer overall survival (OS) and disease-specific survival (DSS). Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Set Enrichment Analysis (GSEA) results showed that ADRB2 participates in immune response. The expression of ADRB2 was positively correlated with the infiltration level of most immune cells. Notably, ADRB2 is involved in LUAD progression partly by regulating the immune microenvironment, which may potentially serve as a significant prognostic biomarker as well as a potential drug target.
Similar content being viewed by others
Introduction
Lung cancer is the primary cause of malignant tumor mortality globally1. LUAD, one of the highest mortality rates and most aggressive forms of cancer, with a low 5-year survival rate < 5%2. Late diagnosis may lead to difficulties in the treatment and prediction of prognosis. Thus, an in-depth study of the molecular mechanisms underlying LUAD progression is urgently needed. At present, there remains an unmet clinical need for tumor biomarkers, and the search for these could lead to more effective treatments and longer survival.
ADRB2 encodes beta-2-adrenergic receptor (β2-AR) which is a member of the G protein-coupled receptor superfamily (GPCRs). GPCRs consist of a large family of integral membrane proteins with seven transmembrane helices. Adrenergic receptors (ARs), a member of GPCRs, are classically divided into two main groups: α-and β-adrenoceptors (β-AR, which is divided into β1, β2, and β3 subtypes)3. β-AR could facilitate cell proliferation, migration, invasion, inflammation, angiogenesis, apoptosis, cell immune response, and epithelial-mesenchymal transition by regulating multiple cancer-related cellular processes. Dysregulated expression of ADRB2 was observed in various cancers, including breast cancer4, hepatocellular carcinoma5, prostate cancer6, and ovarian carcinoma7. Moreover, abundant ADRB2 expression was found to be closely linked with poor clinicopathological characteristics, tumor recurrence, metastasis, and poor prognosis. Although ADRB2 is a carcinogenic biomarker; however, the clinical significance of its expression in patients with LUAD has not been thoroughly elucidated yet.
It is well known that the tumorigenesis, progression, OS, prognosis, and relapse of tumors are strongly linked to the expression of tumor genes. The gene encoding β2-AR, ADRB2, maps to human chromosome 5q31–q32 and is composed of a single exon of 2015 nucleotides8. The effect of ADRB2 on lung cancer remains controversial. Mei et al.9 identified ADRB2 polymorphisms that were correlated with increased lung cancer risk. Nevertheless, Zheng et al.1 found that ADRB2 was underexpressed in LUAD tissues and low ADRB2 expression is associated with poor clinical outcomes. The other research by Wang et al. reached the same conclusion10.
Based on The Cancer Genome Atlas (TCGA) dataset, LUAD dataset was acquired for bioinformatics analysis to verify that ADRB2 expression was significantly down-regulated in LUAD. Next, the relationship between ADRB2 gene expression and clinical traits was further investigated. The expression of ADRB2 was highly correlated with immune infiltration, which further confirmed that ADRB2 could be used as a prognostic biomarker of LUAD.
Results
Patient characteristics
The clinical data of 535 LUAD patients included gender, age, smoking status, T stage, N stage, M stage, pathological stage, OS event, and DSS event (Table 1). Chi-square test revealed that ADRB2 was significantly correlated with T stage (P < 0.001), gender (P = 0.005), smoking (P = 0.004), OS events (P < 0.001) and DSS events (P = 0.005). There was no significant correlation between ADRB2 expression and other clinicopathological features.
ADRB2 expression level in LUAD
Based on the TCGA database, ADRB2 mRNA expression level was analyzed in 594 tissues. As showcased in Fig. 1a, it evaluated the ADRB2 mRNA expression levels in TCGA pan-cancer. The results indicated that the expression level of ADRB2 in the LUAD samples was much higher than that in the normal tissue samples (P < 0.001). Box plots showed ADRB2 mRNA expression levels in 59 adjacent non-tumor tissues and 535 LUAD tissues. As shown in Fig. 1b, ADRB2 was down-expression in LUAD tissues compared with those in normal tissues (P < 0.001, Fig. 1b). Figure 1c shows a pairwise boxplot of the same outcomes (P < 0,001). Figure 1d shows 100 normal samples and 94 LUAD samples from the GEO database, which shows that ADRB2 is lowly expressed in LUAD patients (P < 0,001). ADRB2 expression in LUAD patients does not show correlation to age (Fig. 1e). Moreover, ADRB2 was significantly lower in males (P < 0.001, Fig. 1f) and in patients with a smoking history (P < 0.001, Fig. 1g).
Relative expression level of ADRB2 in LUAD from TCGA database and GEO database. (a) ADRB2 mRNA expression levels in pan-cancers from TCGA database. (b) Boxplot of ADRB2 expression between the LUAD and normal tissues from TCGA database (Normal = 59 and Tumor = 535). (c) Pairwise boxplot of ADRB2 expression between the LUAD and normal tissues from TCGA database (Normal = 57 and Tumor = 57). The differential expression of ADRB2 in GSE40791 from GEO database (Normal = 100 and Tumor = 96) (d).The expression of ADRB2 is grouped by age (e), gender (f), and smoking status (g). ***P < 0.001, ns not significance.
Association between ADRB2 and TNM stages in LUAD patients
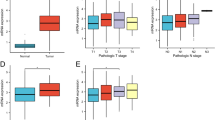
To better understand the impact of ADRB2 on LUAD patient prognosis, Kruskal–Wallis analysis and Spearman correlation analysis were performed to determine the relationship between ADRB2 expression and clinicopathological characteristics (pathologic and TNM stages). The ADRB2 expression was significantly decreased in LUAD patients, and was significantly correlated with pathological stage, T stage and N stage. (Fig. 2a–d, Supplementary Table S1).
Relationship between ADRB2 and clinical characteristics
To further investigate the mechanism of ADRB2 in LUAD, the associations between ADRB2 expression and clinical characteristics were investigated. Based on the clinical data of 535 patients with LUAD, logistic regression analysis indicated that the expression level of ADRB2 in LUAD was negatively correlated with gender (OR 0.603 for males vs. females, P = 0.004); smoking status (OR 0.457 for yes vs. no, P = 0.003); T classification (OR 0.581 for T2 vs. T1, P = 0.005; OR 0.308 for T3 vs. T1, P < 0.001; OR 0.293 for T4 vs. T1, P = 0.018); and pathologic stage (OR 0.527 for stage III vs. stage I, P = 0.011, Fig. 3).
Impact of ADRB2 on the prognosis of LUAD
Survival curves were derived to assess the prognosis of high and low-ADRB2 expression in LUAD patients. As displayed in Fig. 4, patients in the low ADRB2 expression group presented with significantly poorer OS (HR 0.65(0.48–0.87), P = 0.004, Fig. 4a) and DSS (HR 0.61(0.42–0.88), P = 0.008, Fig. 4b) than those in the high ADRB2 expression group. However, PFI did not differ between the two groups (HR 0.90(0.69–1.17), P = 0.412, Fig. 4c). Subgroup analysis suggested that the link between lower ADRB2 expression and worse OS was statistically significant in most subgroups, especially in the stage I subgroup of pathologic stage (HR 0.61(0.38–1), P = 0.048), N0 subgroup of N stage (HR 0.62(0.40–0.94), P = 0.025), M0 subgroup of M stage (HR 0.62(0.44–0.88), P = 0.007).
Effect of ADRB2 expression on survival based on univariate and multivariate analyses
Univariate analysis revealed that pathological stage (HR 2.664; 95% CI 1.960–3.621; P < 0.001); T stage (HR 2.317; 95% CI 1.591–3.375; P < 0.001); N stage (HR 2.601; 95% CI 1.944–3.480; P < 0.001); M stage (HR 2.136; 95% CI 1.248–3.653; P = 0.006); and ARDB2 expression (HR 0.612; 95% CI 0.456–0.821; P = 0.095) were meaningful indicators of survival, and multivariate Cox risk regression analysis showed that T stage (HR 1.817; 95% CI 1.119–2.951; P = 0.016) and N stages (HR 2.086; 95% CI 1.387–3.139; P < 0.001) were independent prognostic factors for OS (Table 2).
Evaluation of the diagnostic capacity of ADRB2 in LUAD
To explore the diagnostic value of ADRB2 for LUAD, receiver operating characteristic (ROC) curve analysis was performed. The results of the ROC curves indicated that ADRB2 was highly sensitive to the diagnosis of LUAD (AUC, 0.994; 95% CI 0.989–0.999, Fig. 5a). Additionally, the AUC was 0.598 for OS, which indicated that the prognostic model had good performance in predicting survival prognosis of patients with LUAD (AUC, 0.590; 95% CI 0.548–0.648, Fig. 5b). The time-dependent accuracy of ADRB2 in predicting OS in 1, 3, and 5 years was also assessed through a time-dependent ROC analysis, AUC < 0.5 indicates that the expression of ADRB2 is opposite to the occurrence trend of OS events in LUAD patients (Fig. 5c). Figure 5d shows that the survival rate of patients in the high-risk group is poor, and the risk of death is high. With the decrease of ADRB2 expression, the risk score tend to increase gradually.
Relationship of ADRB2 expression level with immune infiltration in LUAD
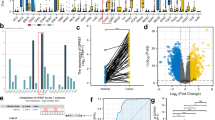
Figure 6a shown the relationships between expression of ADRB2 and 28 types of tumor-infiltrating lymphocytes (TILs) across human cancers in the TISIDB database. In TCGA database, pearson’s analysis demonstrated that the infiltration of 24 types of immune cells was markedly related to ADRB2 expression, which had a significantly positive relationship with CD8 T cells (P = 0.049), type 17 Th cells (Th17) (P = 0.021), and regulatory T cells (TReg) (P = 0.025), and a strongly-positive association with activated DCs (aDCs), B cells, cytotoxic cells, dendritic cells (DCs), eosinophils, immature DCs (iDCs), macrophages, mast cells, neutrophils, natural killer (NK) cells, neutrophils, plasmacytoid DCs (pDCs), T cells, T helper cells, Tcm T central memory (Tcm), T effector memory (Tem), T follicular helper (TFH), and type 1 Th cells (Th1) (P < 0.001, Fig. 6b,c). However, T gamma delta (Tgd) and type 2 Th cells (Th2) (P < 0.001, Fig. 6b,c) showed a negative association with ADRB2. As can be seen in Fig. 6d–i, the expression of ADRB2 was correlated with adundance of B cells (r = 0.334, P = 7.59e−15), CD4 T cells (r = 0.125, P = 0.00448), CD8 T cells (r = 0.27, P = 5.52e−10), macrophage (r = 0.403, P < 2.2e−16), NK cells (r = 0.424, P < 2.2e−16), mast cells (r = 0.527, P < 2.2e−16) in the TISIDB database.
Relations between the expression of ADRB2 and 28 types of TILs across human cancers (a). The relationship between immune cell infiltration and ADRB2 expression (b). The infiltration levels of immune cell populations in lung adenocarcinoma (LUAD) patients with different ADRB2 expression (c). ADRB2 was correlated with abundance of B cells, CD4 T cells, CD8 T cells, macrophage, NK cells, and mast cells (d–i). *P < 0.05, **P < 0.01, ***P < 0.001.
ADRB2 associated gene set enrichment in LUAD
To determine ADRB2-related signaling pathways, GSEA was performed between the high- and low-ADRB2 groups. Significance was assessed using a normalized enrichment score (NES) ≥ 1.5, P ≤ 0.05, and false discovery rate (FDR) ≤ 0.25. KEGG pathway enrichment analysis indicated that 13 important signaling pathways were significantly enriched in the highly expressed ADRB2 phenotypes, including the JAK STAT signaling pathways, leukocyte trans-endothelial migration, chemokine signaling pathway, autoimmune, thyroid disease, Fc epsilon ri signaling pathway, intestinal immune network for iga production, cytokine receptor interaction, B cell receptor signaling pathway, NK cell-mediated cytotoxicity, allograft rejection, Mapk signaling pathway, T cell receptor signaling pathway, and NSCLC. Meanwhile, there were 13 eligible signaling pathways enriched in the low-ADRB2 expression, including spliceosome, RNA polymerase, RNA degradation, citrate cycle (or TCA cycle), cell cycle, pentose phosphate pathway, basal transcription factors, oxidative phosphorylation, DNA replication, mismatch repair, cysteine and methionine metabolism, ubiquitin-mediated-proteolysis, and amino sugar and nucleotide sugar metabolism (Table 3, Fig. 7). These results contribute to further exploration of ADRB2 pathophysiological mechanisms.
Discussion
LUAD is a type of malignant lung tumor that originates from the bronchial mucosal glandular epithelium. LUAD is characterized by inconspicuous early symptoms, and LUAD is a lung tumor with a significant rate of malignant recurrence, metastasis, and unsatisfactory prognosis. Early diagnosis is difficult in most patients.
ADRB2 is ubiquitously expressed in multiple tissues, including the smooth muscle of the human bronchi, cardiovascular system, central nervous system, and gastrointestinal tract. In recent years, increasing evidence shows that ADRB2 has a vital place in the occurrence and development of diverse range of cancers. Zhang et al.11 found that the mRNA expressions of ADRB2 were higher in gastric cancers compared with normal tissues. Moreover, patients with gastric cancer with positive ADRB2 expression exhibited larger tumor size, late clinical stage, lower differentiation, and distant metastasis. In addition, high ADRB2 expression can promote the angiogenic switch in prostate cancer and prevent or delay the dominant role of pro-angiogenic factors, leading to tumor progression12. β2-AR is encoded by ADRB2 and can bind specifically to endogenous catecholamines (such as adrenaline and noradrenaline), and promotes the production and release of cyclic adenosine phosphate (cAMP). cAMP can further activate and phosphorylate protein kinase A and C to activate downstream signal transduction pathways and promote the proliferation, migration, and metastasis of lung cancer cells13. The positive ADRB2 expression can occur in several cancers, including hepatocellular carcinoma, colorectal cancer, melanoma, and gastric cancer, and is often indicative of poor prognosis5,11,14,15,16. The regulation of β2-AR on tumorigenesis may be twofold, depending on the type of tumor and the stage of cancer progression. A study have revealed that activation of ADRB2 results in inhibition tumor cell growth, as well as induction of apoptosis and tumor regression, where this activation results in the inactivation of the Raf-1/Mek-1/Erk1/2 pathway by a cAMP-dependent activation of protein kinase A17. In oral squamous cell carcinoma, patients with higher ADRB2 had a significant longer DSS and OS18. Caparica et al.19 found that a high ADRB2 expression may be a favorable prognostic factor in patients with HER2 + early breast cancer and evidence that a possible association with antiproliferative, antiangiogenic, and immunogenic effects of ADRB2. Yazawa et al.20 retrospectively analyzed 328 surgically-resected patients with NSCLC and found that positive ADRB2 expression was found in 29% of LUAD tissues, which markedly increased compared with in non-adenocarcinoma tissues. A high level of ADRB2 expression was associated with vascular invasion, tumor cell proliferation, and poor prognosis in patients with LUAD. Nevertheless, Wang et al.10 searched the gene expression synthesis (GEO) to obtain data showing that ADRB2 is down-regulated in LUAD, and ADRB2 mRNA levels declined with stage progression. ADRB2 mRNA expression levels and its gene product, β2-AR, differ. The exact mechanism is still controversial. This study used high-throughput RNA sequencing data obtained from TCGA database to further ascertain the expression and the prognostic significance of ADRB2 in LUAD and explore its correlation with immune cell infiltration.
The low expression of ADRB2 in tumor tissues of LUAD patients is correlated with gender, smoking, OS events and DSS events, indicating that ADRB2 is a potential biomarker for prognosis and diagnosis. Data mining from TCGA database showed that the expression of ADRB2 was correlated with pathological stage, T stage and N stage, and the disease tended to progress with the decrease of ADRB2 expression. Premised on this, logistic regression analysis showed that ADRB2 is significantly correlated with gender, smoking status, T stage and pathological stage. The survival analysis revealed a favorable survival in high ADRB2 expression group compared to those with low expression. Univariate analysis revealed that pathological stage, T stage, N stage, M stage, and ADRB2 expression influenced OS. To further determine the diagnostic capacity of ADRB2 in LUAD, ROC curves were used to confirm that ADRB2 is sensitive to the diagnosis and prognosis of LUAD. Altogether, these findings illustrate that ADRB2 is a potential prognostic biomarker for LUAD.
The tumor immune microenvironment (TME) is very important in cancer pathogenesis21. Immune cells are vital elements of the TME22. The correlation between ADRB2 expression and the infiltration of 24 immunocytes was further explored to elucidate the mechanisms responsible for ADRB2 to predict clinical prognosis. Further correlation analysis indicated that the infiltration of 19 immune cells was significantly associated with ADRB2 expression. ADRB2 expression was positively correlated with aDCs, B cells, CD8+ T cells, cytotoxic cells, DCs, eosinophils, iDCs, macrophages, mast cells, neutrophils, NK cells, pDCs, T cells, T helper cells, Tcm, Tem, TFH, Th1 cells, Th17 cells, and TReg. B cells are dominant in the progression of lung cancers23,24 and can be observed at the individual stages of carcinogenesis25. B cells can prolong the survival of cancer patients by inhibiting tumor progression and preventing metastasis. In addition, antibodies produced by B cells are essential mediators of tumor cell death26. Cytotoxic T lymphocytes (CTLs) are major players in antitumor immunity and can lead to apoptosis of cancer cells through a series of steps; therefore, high infiltration of CTLs is a favorable prognostic marker for many cancers. DCs are the most effective antigen-presenting cells to induce primary tumor immune response27, and in NSCLC patients, an increased DC count was significantly associated with an increase in DSS28. The role of macrophages in cancer progression is still controversial. Tumor-associated macrophages promote tumor progression by facilitating tumor stroma formation and angiogenesis29. In patients with NSCLC with prolonged survival, macrophage-infiltrating tumors are mainly of the M1 type30. Mast cells, which have cytotoxic effects on cancer cells, can enhance the immunity of patients with LUAD against cancer cells and improve their postoperative prognosis31. NK cells are cytotoxic and it is essential in the immune monitoring of cancers32. Carrega et al.33 found that in resected LUAD tissues, increasing numbers of infiltrating NK cells were associated with favorable patient survival outcomes. T cells are the most abundant monocytes infiltrating the NSCLCs34. T cells can secrete cytokines to inhibit tumor stroma formation and use cytotoxic molecules to kill epithelial nuclear stromal cells. Al-Shibli et al.35 reported that T cell infiltration is associated with better DSS, and T cells are an independent indicator of survival. T helper cells play an important role in cancer immunity by secreting cytokines36. Both Th1 and Th17 cells produce proinflammatory factors, and their extensive infiltration can significantly improve clinical outcomes in a variety of cancers37,38. Large infiltration of cytotoxic T cells in tumor tissues is associated with longer survival35. Studies have shown that TFH has an antitumor response, and IL-21 secreted by TFH induces the activation, proliferation, and differentiation of B cells39,40. ADRB2 expression may up-regulate the levels of infiltrating immune cells to limit the development of LUAD. In contrast, ADRB2 expression was negatively correlated with Th2 cells and Tgd. Th2 cells have many pro-neoplastic activities and take part in cancer progression by cytokine release. Th2 cells are dominant in lymphocytes from malignant pleural effusion in patients with lung cancer41,42. Current studies have revealed that Tgd has a pro-tumor effect, which can inhibit innate and adaptive immunity by inducing immunosenescence43,44,45.
To summarize, low ADRB2 expression is associated with poor prognosis of LUAD. These results show that ADRB2 expression level affects the immunity activity in the TME, and ADRB2 might be a valuable biomarker for the immune status in LUAD patients.
To further explore the mechanism of ADRB2 in LUAD, the signaling pathways involved in ADRB2 was screened. In the ADRB2 high-expression group, ADRB2 associated genes were significantly enriched in immune signaling pathways (such as B cell receptor signaling pathway, T cell receptor signaling pathway, and NSCLC, NK-cell-mediated cytotoxicity, chemokine signaling pathway, and Jak STAT signaling pathway), in KEGG analysis. Those are significant in the tumorigenesis, development, and invasion of malignancies46. On the other hand, in the ADRB2 low-expression group, ADRB2 correlated genes were enriched in metabolism-related pathways, including RNA polymerase, citrate cycle, pentose phosphate pathway, oxidative phosphorylation, cysteine and methionine metabolism, and amino sugar and nucleotide sugar metabolism, implying that ADRB2 up-regulated the signaling pathways associated with immune response and induced antitumor efficiency. Therefore, ADRB2 expression was down-regulated as LUAD progressed, and the TME switched from an immune-active state to a metabolic state. The ADRB2 expression can be considered a biomarker to predict immune response.
At present, most studies on the relationship between ADRB2 and the occurrence and progression of LUAD are based on its gene expression product, β2-AR, and its signaling pathway. This study revealed ADRB2 as a key gene in the immune microenvironment of LUAD by performing a bioinformatics analysis, to provide evidence for ADRB2 as a potential prognostic marker for LUAD. However, some limitations arise in the research. Firstly, the present research was limited by the small number of cases, and a large cohort is needed to validate the results of this research. Secondly, this research primarily focused on the expression of ADRB2 mRNA from TCGA, and without involving β2-AR levels in LUAD tissues. Thus, this study still needs large-sample, multi-center, multi-ethnic clinical trials and basic experimental studies to prove the prognostic value of ADRB2 in LUAD. Low expression of ADRB2 is associated with poor prognosis in LUAD patients, which may be related to immunocompromised.
Conclusion
In summary, ADRB2 expression was significantly down-regulated in patients with LUAD. ADRB2 is involved in LUAD progression partly by regulating the immune microenvironment, which may potentially serve as a significant prognostic biomarker as well as a potential drug target.
Materials and methods
Data acquisition
On or before November 13, 2021, the mRNA profile (HTSeq-Counts and HTSeq-FPKM) was extracted from TCGA (https://cancergenome.nih.gov/), including 535 LUAD samples and 59 normal samples. Relevant clinical information was derived from TCGA. RNAseq data in FPKM (Fregments Per Kilobase Per Million) format were converted to TPM (transcripts Per Million reads) format and log2 transformed. ADRB2 expression data from datasets GSE40791 was downloaded from the GEO database (https://www.ncbi.nlm.nih.gov/gds/). The relevant data TCGA and GEO provided is open-access, no additional approval from the Ethics Committee were required. All methods were performed in accordance with the relevant guidelines and regulations.
ADRB2 expression and survival analyses
The original expression data downloaded from TCGA were processed using the Perl programming language (version: strawberry-perl-5.32.1.1-64bit.mis, https://strawberryperl.com/). GEOquery package (version: 2.54.1, https://cran.r-project.org/web/packages/GEOquery/index.html) is used to download GEO database expression data. The differential ADRB2 expression were analyzed by Mann–Whitney U test, Dunn’s test, Kruskal–Wallis test or Spearman correlation analysis when appropriate, and the results were visualized using the ggplot2 R package (version: 3.3.2, https://cran.r-project.org/web/packages/ggplot2/index.html). Binary logistics regression model was used to evaluate the relationship between ADRB2 and clinical characteristics. Survival data were extracted and analyzed using the Perl programming language, and patients without complete survival state and time were removed. Subsequently, we matched the complete survival data with ADRB2 expression data and obtained 499 patients’ data. In survival analysis, the ADRB2 mRNA expression level was split into two groups by the median expression value, and OS, DSS, and progression-free interval (PFI) were evaluated with cox regression and log-rank test. A Kaplan–Meier survival curve was constructed by the survival (version:3.2-10 https://cran.r-project.org/web/packages/survivalAnalysis/index.html) and survmine (version:0.4.9,https://cran.r-project.org/web/packages/survminer/index.html) package of R software. pROC R package (version: 1.17.0.1, https://cran.r-project.org/web/packages/pROC/index.html) and timeROC R package (version:0.4,https://cran.r-project.org/web/packages/timeROC/index.html) were used for statistical analysis, and ggplot2 R package was used for visualization when drawing ROC curves and risk score map.
Univariate and multivariate Cox regression analyses
Both univariate and multivariate analyses of clinical pathological parameters were performed adopting Cox proportional hazards analysis. In addition, we quantitatively evaluated the independent predictive value of clinicopathological parameters and ADRB2 expression for survival and explored the prognostic effect of ADRB2 on survival after adjusting for other confounding factors. Meanwhile, when matching it with ADRB2 expression data, incomplete clinical information was excluded.
Evaluation of immune infiltration
Figure 6a is drawn online in TISIDB (http://cis.hku.hk/TISIDB/index.php). GSEA method from the R package “GSVA (version:1.34.0, http://bioconductor.org/packages/release/bioc/html/GSVA.html), clusterProfiler (version:3.18.0, http://bioconductor.org/packages/release/bioc/html/clusterProfiler.html), and rtracklayer (http://bioconductor.org/packages/release/bioc/html/rtracklayer.html)” was used to present infiltration enrichment of 24 common immune cells in each sample, including mast cells, DCs, iDCs, macrophages, eosinophils, TFH, Th1, neutrophils, pDCs, T cells, NK cells, B cells, aDCs, Tem, T helper cells, cytotoxic cells, Tcm, CD8 T cells, TReg, NK CD56 bright cells, Th17, NK CD56dim cells, Tgd, and Th2. After that, Pearson’s analysis was used to investigate the relationship between ADRB2 expression level and 24 immune cell infiltration in LUAD. Pearson correlation test and the independent-samples T test were used to compare the levels of immune cell infiltration between different ADRB2 expression groups. Spearman’ test was used to evaluate the relationship between ADRB2 expression and the abundance of TILs.
Gene set enrichment analysis (GSEA)
All LUAD patients in TCGA dataset were allocated into high and low group based on the expression of ADRB2. GSEA was used as a signaling pathway analysis tool to explore the signaling pathways related to ADRB2 in LUAD. GSEA between high and low ADRB2 expression was performed using GSEA 3.0 (http://www.gsea-msigdb.org/gsea/index.jsp). Phenotypes were determined based on ADRB2 expression levels. The gene set “c2 all.v6.0 symbols.gmt” was used for the enrichment analysis. KEGG analysis was performed to explore the significant pathways associated with ADRB2 expression.
Statistical analysis
All statistical analyses were performed using IBM SPSS Statistics for Windows, version 23.0 (IBM Corp, Armonk, NY, USA) and R (version 4.0.2, https://www.r-project.org/) and the level of statistical significance was defined as a P < 0.05.
Data availability
The datasets generated and/or analysed during the current study are available in TCGA dataset (https://cancergenome.nih.gov/) and GEO database (https://www.ncbi.nlm.nih.gov/gds/).
References
Zheng, Q., Min, S. & Zhou, Q. Identification of potential diagnostic and prognostic biomarkers for LUAD based on TCGA and GEO databases. Biosci. Rep. https://doi.org/10.1042/bsr20204370 (2021).
Lu, X. et al. Phosphoinositide specific phospholipase Cgamma1 inhibition-driven autophagy caused cell death in human lung adenocarcinoma A549 cells in vivo and in vitro. Int. J. Biol. Sci. 16, 1427–1440. https://doi.org/10.7150/ijbs.42962 (2020).
Ruffolo, R. R. Jr. & Hieble, J. P. Alpha-adrenoceptors. Pharmacol. Ther. 61, 1–64. https://doi.org/10.1016/0163-7258(94)90058-2 (1994).
Qin, J. F. et al. Adrenergic receptor beta2 activation by stress promotes breast cancer progression through macrophages M2 polarization in tumor microenvironment. BMB Rep. 48, 295–300. https://doi.org/10.5483/bmbrep.2015.48.5.008 (2015).
Chen, D. et al. The beta2-adrenergic receptor is a potential prognostic biomarker for human hepatocellular carcinoma after curative resection. Ann. Surg. Oncol. 19, 3556–3565. https://doi.org/10.1245/s10434-012-2396-1 (2012).
Ramberg, H. et al. Hormonal regulation of beta2-adrenergic receptor level in prostate cancer. Prostate 68, 1133–1142. https://doi.org/10.1002/pros.20778 (2008).
Thaker, P. H. et al. Chronic stress promotes tumor growth and angiogenesis in a mouse model of ovarian carcinoma. Nat. Med. 12, 939–944. https://doi.org/10.1038/nm1447 (2006).
Litonjua, A. A. et al. Very important pharmacogene summary ADRB2. Pharmacogenet. Genom. 20, 64–69. https://doi.org/10.1097/FPC.0b013e328333dae6 (2010).
Mei, L., Huang, C., Wang, A. & Zhang, X. Association between ADRB2, IL33, and IL2RB gene polymorphisms and lung cancer risk in a Chinese Han population. Int. Immunopharmacol. 77, 105930. https://doi.org/10.1016/j.intimp.2019.105930 (2019).
Wang, Z. et al. Decreased HLF expression predicts poor survival in lung adenocarcinoma. Med. Sci. Monit. 27, e929333. https://doi.org/10.12659/MSM.929333 (2021).
Zhang, X. et al. Chronic stress promotes gastric cancer progression and metastasis: An essential role for ADRB2. Cell Death Dis. 10, 788. https://doi.org/10.1038/s41419-019-2030-2 (2019).
Zahalka, A. H. et al. Adrenergic nerves activate an angio-metabolic switch in prostate cancer. Science 358, 321–326. https://doi.org/10.1126/science.aah5072 (2017).
Huang, Q. et al. The role of adrenergic receptors in lung cancer. Am. J. Cancer Res. 8, 2227–2237 (2018).
Shimizu, A. et al. Prognostic significance of beta2-adrenergic receptor expression in malignant melanoma. Tumour Biol. 37, 5971–5978. https://doi.org/10.1007/s13277-015-4420-0 (2016).
Takahashi, K. et al. Clinical significance of beta2-adrenergic receptor expression in patients with surgically resected gastric adenocarcinoma. Tumour Biol. 37, 13885–13892. https://doi.org/10.1007/s13277-016-5139-2 (2016).
Florescu, C. et al. The evaluation of Beta-2-adrenoreceptors’ expression in normal peritumoral tissue in patients with colorectal adenocarcinoma. Curr. Health Sci. J. 42, 335–341. https://doi.org/10.12865/chsj.42.04.01 (2016).
Carie, A. E. et al. A chemical biology approach identifies a beta-2 adrenergic receptor agonist that causes human tumor regression by blocking the Raf-1/Mek-1/Erk1/2 pathway. Oncogene 26, 3777–88. https://doi.org/10.1038/sj.onc.1210172 (2007).
Bravo-Calderon, D. M. et al. Beta adrenergic receptor activation inhibits oral cancer migration and invasiveness. Arch. Oral Biol. 118, 104865. https://doi.org/10.1016/j.archoralbio.2020.104865 (2020).
Rafael, C. et al. Prognostic and predictive impact of Beta-2 adrenergic receptor expression in HER2-positive breast cancer. Clin. Breast Cancer 20, 262–273 (2020).
Yazawa, T. et al. Prognostic significance of β2-adrenergic receptor expression in non-small cell lung cancer. Am. J. Transl. Res. 8, 5059–5070 (2016).
Schulz, M., Salamero-Boix, A., Niesel, K., Alekseeva, T. & Sevenich, L. Microenvironmental regulation of tumor progression and therapeutic response in brain metastasis. Front. Immunol. 10, 1713. https://doi.org/10.3389/fimmu.2019.01713 (2019).
DeNardo, D. G. & Ruffell, B. Macrophages as regulators of tumour immunity and immunotherapy. Nat. Rev. Immunol. 19, 369–382. https://doi.org/10.1038/s41577-019-0127-6 (2019).
Banat, G. A. et al. Immune and inflammatory cell composition of human lung cancer stroma. PLoS ONE 10, e0139073. https://doi.org/10.1371/journal.pone.0139073 (2015).
Kurebayashi, Y. et al. Comprehensive immune profiling of lung adenocarcinomas reveals four immunosubtypes with plasma cell subtype A negative indicator. Cancer Immunol. Res. 4, 234–247. https://doi.org/10.1158/2326-6066.Cir-15-0214 (2016).
Bian, T. et al. Prognostic biomarker TUBA1C is correlated to immune cell infiltration in the tumor microenvironment of lung adenocarcinoma. Cancer Cell Int. 21, 144. https://doi.org/10.1186/s12935-021-01849-4 (2021).
Bremnes, R. M. et al. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: Emphasis on non-small cell lung cancer. J. Thorac. Oncol. 6, 824–833. https://doi.org/10.1097/JTO.0b013e3182037b76 (2011).
Wang, Y. et al. The role of ADRB2 gene polymorphisms in malignancies. Mol. Biol. Rep. 48, 2741–2749. https://doi.org/10.1007/s11033-021-06250-y (2021).
McGraw, D. W. et al. Polymorphisms of the 5’ leader cistron of the human beta2-adrenergic receptor regulate receptor expression. J. Clin. Investig. 102, 1927–1932. https://doi.org/10.1172/jci4862 (1998).
de Visser, K. E., Eichten, A. & Coussens, L. M. Paradoxical roles of the immune system during cancer development. Nat. Rev. Cancer 6, 24–37. https://doi.org/10.1038/nrc1782 (2006).
Ma, J. et al. The M1 form of tumor-associated macrophages in non-small cell lung cancer is positively associated with survival time. BMC Cancer 10, 112. https://doi.org/10.1186/1471-2407-10-112 (2010).
Tomita, M., Matsuzaki, Y. & Onitsuka, T. Correlation between mast cells and survival rates in patients with pulmonary adenocarcinoma. Lung Cancer 26, 103–108. https://doi.org/10.1016/s0169-5002(99)00076-8 (1999).
Waldhauer, I. & Steinle, A. NK cells and cancer immunosurveillance. Oncogene 27, 5932–5943. https://doi.org/10.1038/onc.2008.267 (2008).
Carrega, P. et al. Natural killer cells infiltrating human nonsmall-cell lung cancer are enriched in CD56 bright CD16(-) cells and display an impaired capability to kill tumor cells. Cancer 112, 863–875. https://doi.org/10.1002/cncr.23239 (2008).
Kataki, A. et al. Tumor infiltrating lymphocytes and macrophages have a potential dual role in lung cancer by supporting both host-defense and tumor progression. J. Lab. Clin. Med. 140, 320–328. https://doi.org/10.1067/mlc.2002.128317 (2002).
Al-Shibli, K. et al. The prognostic value of intraepithelial and stromal CD3-, CD117- and CD138-positive cells in non-small cell lung carcinoma. APMIS 118, 371–382. https://doi.org/10.1111/j.1600-0463.2010.02609.x (2010).
Katsuta, E., Rashid, O. M. & Takabe, K. Clinical relevance of tumor microenvironment: Immune cells, vessels, and mouse models. Hum Cell 33, 930–937. https://doi.org/10.1007/s13577-020-00380-4 (2020).
Aldarouish, M. et al. Immunomodulatory effects of chemotherapy on blood lymphocytes and survival of patients with advanced non-small cell lung cancer. Int. J. Immunopathol. Pharmacol. 33, 2058738419839592. https://doi.org/10.1177/2058738419839592 (2019).
Song, L. et al. Long-term prognostic significance of interleukin-17-producing T cells in patients with non-small cell lung cancer. Cancer Sci. 110, 2100–2109. https://doi.org/10.1111/cas.14068 (2019).
Kang, S. G. et al. MicroRNAs of the miR-17∼92 family are critical regulators of T(FH) differentiation. Nat. Immunol. 14, 849–857. https://doi.org/10.1038/ni.2648 (2013).
Lu, L. et al. Repurposing of miltefosine as an adjuvant for influenza vaccine. Vaccines https://doi.org/10.3390/vaccines8040754 (2020).
Zhu, J., Yamane, H. & Paul, W. E. Differentiation of effector CD4 T cell populations (*). Annu. Rev. Immunol. 28, 445–489. https://doi.org/10.1146/annurev-immunol-030409-101212 (2010).
Oshikawa, K., Yanagisawa, K., Ohno, S., Tominaga, S. & Sugiyama, Y. Expression of ST2 in helper T lymphocytes of malignant pleural effusions. Am. J. Respir. Crit. Care Med. 165, 1005–1009. https://doi.org/10.1164/ajrccm.165.7.2105109 (2002).
Fonseca, S. et al. Human peripheral blood gamma Delta T cells: Report on a series of healthy Caucasian Portuguese adults and comprehensive review of the literature. Cells https://doi.org/10.3390/cells9030729 (2020).
Ye, J. et al. Tumor-derived γδ regulatory T cells suppress innate and adaptive immunity through the induction of immunosenescence. J. Immunol. 190, 2403–2414. https://doi.org/10.4049/jimmunol.1202369 (2013).
Peng, G. et al. Tumor-infiltrating gammadelta T cells suppress T and dendritic cell function via mechanisms controlled by a unique toll-like receptor signaling pathway. Immunity 27, 334–348. https://doi.org/10.1016/j.immuni.2007.05.020 (2007).
Yang, X., Tang, Z., Zhang, P. & Zhang, L. Research advances of JAK/STAT signaling pathway in lung cancer. Zhongguo fei ai za zhi Chin. J. Lung Cancer 22, 45–51. https://doi.org/10.3779/j.issn.1009-3419.2019.01.09 (2019).
Funding
The present study was supported by the National Natural Science Foundation of China (Grant No. 82004281), the China Postdoctoral Science Foundation (Grant Nos. 2021T140427 and 2021M691986), and Young Elite Scientists Sponsorship Program by CAST (Grant No.2021-QNRC2-B25).
Author information
Authors and Affiliations
Contributions
L.Y.J. wrote the original draft, F.X. and S.T. prepared the figures and tables, J.T.Z. and W.D.Ch. analyzed the raw data, X.Y. and X.B.C. downloaded the raw data from TCGA database, X.L. and M.H.G. reviewed the relevant literature, Q.Q.W. made contribution to the language, Z.T.C. edited the manuscript and made revisions. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ji, L., Xu, F., Zhang, J. et al. ADRB2 expression predicts the clinical outcomes and is associated with immune cells infiltration in lung adenocarcinoma. Sci Rep 12, 15994 (2022). https://doi.org/10.1038/s41598-022-19991-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-19991-y
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.