Abstract
Cervical cancer, caused by human papillomavirus (HPV), is the fourth most common type of cancer among women worldwide. While HPV prophylactic vaccines are available, they have no therapeutic effects and do not clear up existing infections. This study aims to design a therapeutic vaccine against cervical cancer using reverse vaccinology. In this study, the E6 and E7 oncoproteins from HPV16 were chosen as the target antigens for epitope prediction. Cytotoxic T lymphocytes (CTL) and helper T lymphocytes (HTL) epitopes were predicted, and the best epitopes were selected based on antigenicity, allergenicity, and toxicity. The final vaccine construct was composed of the selected epitopes, along with the appropriate adjuvant and linkers. The multi-epitope vaccine was evaluated in terms of physicochemical properties, antigenicity, and allergenicity. The tertiary structure of the vaccine construct was predicted. Furthermore, several analyses were also carried out, including molecular docking, molecular dynamics (MD) simulation, and in silico cloning of the vaccine construct. The results showed that the final proposed vaccine could be considered an effective therapeutic vaccine for HPV; however, in vitro and in vivo experiments are required to validate the efficacy of this vaccine candidate.
Similar content being viewed by others
Introduction
Cervical cancer, with about 0.6 million cases and 0.3 million deaths per year, is the fourth most common type of cancer among women worldwide1. Human papillomavirus (HPV) is the most important cause of this disease, which is transmitted through sexual intercourse2. There are five main genera of HPV, alpha, beta, gamma, mu, and nu3. The most important HPVs are in the alpha genus, and they are classified into high-risk and low-risk groups based on the risk of oncogenic transformation4. The high-risk group includes types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, and 685. Based on biological studies, HPV16 and 18 together cause approximately 70% of all cervical cancers6, and HPV16 is the most carcinogenic of the two7. HPV types 6, 11, 40, 42, 43, 44, 54, 61, 70, 72, and 81 are classified in the low-risk group8. HPV is a non-enveloped virus with a circular double-stranded DNA genome that is approximately 8 kb in length9. There are three regions in the HPV genome, an early region (E1, E2, E4, E5, E6, and E7), a late region (L1, L2), and a long control region (LCR)10.
The E6 and E7 oncoproteins are the major virus transforming proteins in high-risk HPV, and they play a role in cell proliferation, immortalization, and transformation in human epithelial cells11. The key function of the E6 protein in high-risk HPV types is to promote ubiquitin-mediated degradation of the p53 protein through its interaction with the E6-associated protein (E6AP)12. Moreover, p53 is a transcription factor that regulates the expression of genes involved in cell cycle arrest and apoptosis12. E7 binds to the retinoblastoma (Rb) protein, causing E2F to be released from the Rb-E2F complex and the cell to enter the S phase13. Since the E6 and E7 oncoproteins are essential for tumor progression, and they are consistently expressed in HPV-infected cells but not in healthy cells, they are ideal targets for the development of therapeutic HPV vaccines14,15.
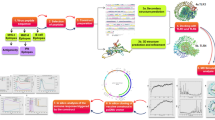
Gardasil and Cervarix are available prophylactic vaccines to prevent HPV infection. These vaccines have no therapeutic effect because their action mechanism is to induce the production of neutralizing antibodies against the L1 capsid protein, and since L1 is expressed in the granular epithelium before viral shedding, consequently, current prophylactic vaccines are not effective in eliminating previous infections16. Inducing cell-mediated immune response is needed to clear infected cells17. This study aims to design a therapeutic vaccine against cervical cancer using reverse vaccinology. The reverse vaccinology method, which examines the genomes of pathogenic microorganisms to identify antigens, employs a number of algorithms for predicting T-cell and B-cell epitopes. Unlike the conventional vaccinology method, this method does not require culturing pathogens and extracting antigenic proteins, which are costly and time-consuming processes18. In the present study, CTL and HTL epitopes of the E6 and E7 oncoproteins were identified and linked together by appropriate linkers for the design of a multi-epitope vaccine against cervical cancer. Multi-epitope vaccines are recombinant vaccines that are considered to be a promising strategy against tumors and viral infections due to their high specificity, safety, and stability, and low-cost development19. The major downside of multi-epitope vaccines is their low immunogenicity because proteinases can quickly degrade the antigenic peptides in the body, making them difficult to identify by the immune cells’ receptors20. One of the strategies suggested for improving the immune response generated by multi-epitope vaccines is to use adjuvants in the vaccine construct21. We also added the 50S ribosomal protein L7/L12 (Locus RL7_MYCTU) as an adjuvant to the N-terminal of the vaccine construct with the help of an EAAAK linker. Subsequently, the physicochemical properties and the secondary and tertiary structures of the vaccine were predicted. Furthermore, several analyses were also carried out, including molecular docking, MD simulation, and in silico cloning of the vaccine construct. The flow of methods used to design a multi-epitope vaccine is illustrated in Fig. 1.
Results
Identification and selection of T-cell epitopes
A total of 54 CTL epitopes for the E6 and E7 proteins were predicted using the NetCTL 1.2 server. The predicted epitopes were screened at several stages. In the first step, those epitopes were selected that could bind to at least three MHC class I supertypes. These epitopes were then evaluated for antigenicity, toxicity, and allergenicity using VaxiJen v2.0, ToxinPred, and AllerTOP v. 2.0 servers, respectively. Finally, a CTL epitope was selected for each of the E6 and E7 proteins (Table 1). Here, we predicted 99 HTL epitopes for the E6 and E7 proteins using the NetMHCII 2.3 server, among which 27 epitopes capable of binding to at least three MHC class II alleles were identified and checked for antigenicity, toxicity, and allergenicity. The selection of six HTL epitopes for E6 and one epitope for E7 was the result of these screenings (Table 2).
Construction of the multi-epitope vaccine construct
A total of two CTL epitopes and seven HTL epitopes were merged to construct the multi-epitope vaccine using AAY and GPGPG linkers, respectively. A sequence of 159 amino acids was generated after epitope fusion. The adjuvant sequence, with a length of 130 amino acid (MAKLSTDELLDAFKEMTLLELSDFVKKFEETFEVTAAAPVAVAAAGAAPAGAAVEAAEEQSEFDVILEAAGDKKIGVIKVVREIVSGLGLKEAKDLVDGAPKPLLEKVAKEAADEAKAKLEAAGATVTVK), was added to the N-terminal of the vaccine sequence by an EAAAK linker. The final designed vaccine construct consisted of 294 amino acids (Fig. 2).
Evaluation of the antigenicity, allergenicity, and physicochemical properties of the vaccine
The antigenicity of the vaccine construct was calculated using the VaxiJen v2.0 and ANTIGENpro servers. The probability of antigenicity predicted by VaxiJen v2.0 and ANTIGENpro was 0.5058 and 0.745186, respectively. The allergenicity of the proposed vaccine was predicted using the AllerTOP v. 2.0 server, indicating that it was non-allergenic. Various physicochemical characteristics of the designed vaccine were calculated using the ProtParam server. The final composition of the multi-epitope vaccine consists of 294 amino acids. The theoretical pI, molecular weight, and instability index of the vaccine construct were calculated to be 8.33, 32.01 kDa, and 37.82, respectively. The half-life of the vaccine was estimated to be 30 h in mammalian reticulocytes, more than 20 h in yeast, and more than 10 h in E. coli. The aliphatic index of the vaccine was 76.43, and its GRAVY score was reported to be -0.307.
Prediction of the secondary structure
The percentage of the secondary structure components of the multi-epitope vaccine was computed using the Prabi server. The predicted structure included alpha-helix (40.48%), extended strand (27.55%), and random coil (31.97%) (Fig. 3).
Tertiary structure modeling, refinement, and validation of the multi-epitope vaccine
The five models of the 3D structure of the vaccine construct were generated by the I-TASSER server using the threading templates (PDB Hit: 1dd4A, 6slmA, 1rquA, 4giz, 1dd3A, 4gizC, 2ftc, 1dd4A, and 6slm). The calculated C-score values for models 1–5 were − 3.72, − 3.94, − 4.10, − 4.43, and − 4.44, respectively. The C-score is usually within the range of − 5 to 2, where a higher C-score for the model indicates that it has a high level of confidence22. Therefore, we selected model 1 with a C-score value of − 3.72. Chimera 1.15rc software was used to visualize the 3D model of the vaccine construct23 (Fig. 4). This model was then refined by the 3Drefine server. This server provided five refined models with different parameters, including the 3D refined score, GDT-TS, GDTHA, RMSD, MolProbity, and RWPlus (Table 3). Higher GDT-TS, GDT-HA, and RMSD values, and lower 3D refine Score, RWplus, and MolProbity values indicate a higher quality for the models. The refined model 5 was selected based on the above parameters (Fig. 4). The ProSA-web and SAVES v6.0 servers were also used to compare the overall quality of the protein structure of the multi-epitope vaccine before and after the refining process. The Z-score of the initial and refined models was − 0.86 and − 2.48, respectively (Fig. 5A,B). The Ramachandran plot generated by the SAVES v6.0 server showed that in the initial model, 50.2%, 34.3%, 10.6%, and 4.9% of the residues were present in the favoured, additional allowed, generously-allowed, and disallowed regions, respectively Fig. 5C), while in the refined model, these values changed to 60.8%, 25.3%, 8.6% and 5.3%, respectively (Fig. 5D).
Evaluation of the 3D model of the vaccine construct using the ProSA-web and Ramachandran plot before and after the refining. (A) The initial model has a Z score of − 0.86, (B) while the refined model has a Z score of − 2.48. (C) The Ramachandran plot analysis shows that in the initial model, 50.2%, 34.3%, 10.6%, and 4.9% of the residues are found in the favoured, additional allowed, generously allowed, and disallowed regions, respectively, (D) while in the refined model, these values changed to 60.8%, 25.3%, 8.6% and 5.3%, respectively.
Prediction of the B-cell epitope
Seven linear B-cell epitopes (20-mer) were predicted by the BCPREDS, and the scores of the epitopes ranged from 0.819 to 1 (Table 4). The position of the linear B-cell in the final vaccine construct were highlighted using the Chimera 1.15rc software23 (Fig. 6). The ElliPro server also predicted five discontinuous B-cell epitopes in the tertiary structure of the vaccine (Fig. 7). The minimum and maximum scores for the predicted discontinuous B-cell epitopes were 0.648 and 0.792, respectively (Table 5).

source molecular viewer Jmol (http://jmol.sourceforge.net/) was employed. The gray sticks and the yellow surface indicate the vaccine construct and discontinuous B-cell epitopes, respectively.
To visualize discontinuous epitopes on the vaccine construct's 3D structure (A–E), the open-
Molecular docking
The molecular docking between the vaccine construct and TLR4 was conducted using the ClusPro 2.0 server. In this study, the server generated 26 clusters, and it then ranked them by energy level. The cluster with the lowest energy of -1103.8 was chosen as the best complex. The Chimera 1.15rc software was used to visualize the molecular docking results23 (Fig. 8). There were 17 hydrogen bonds between chain B of TLR4 and the vaccine, while three hydrogen bonds formed between chain D and the vaccine. The map of hydrogen bonds and hydrophobic contacts between the vaccine construct and TLR4 generated by the LigPlot v1.4.5 program24 (Fig. 9). Tables 6 and 7 show the amino acids involved in the formation of these hydrogen bonds along with the lengths of the bonds.
MD simulation
The docked complex of the vaccine construct and TLR4 was subjected to MD simulation using the GROMACS 2019.6 software. RMSD was evaluated to determine the structural stability of the vaccine and TLR4, while RMSF was calculated to measure residual fluctuations. At the beginning of the simulation, the RMSD value of TLR4 increased rapidly, reaching approximately 0.3 nm in 3000 ps, and it then fluctuated slightly around this value until the end of the simulation. The RMSD value of the vaccine had an upward trend, reaching 0.6 nm at 1000 ps. Afterward, RMSD increased slowly until it reached approximately 1.1 nm at 30,000 ps, and it remained constant at this value until the end of the simulation (Fig. 10A). In the previous section, it was observed that the vaccine construct was attached to chain B from TLR4, and since both chains A and B from TLR4 have the same sequence, the RMSF values of the two chains were compared to accurately evaluate the effects of the vaccine construct binding on the flexibility of chain B. The RMSF value of residues 30–110, 200–205, 390–470, and 550–627 from chain A showed a greater degree of flexibility than that of chain B, while the flexibility of other regions in the two chains was almost the same. The RMSF plot of the vaccine showed that most of the residues were highly flexible (Fig. 10B).
Codon optimization and in silico cloning of the final vaccine construct
The back translation and codon optimization of the multi-epitope vaccine were performed by JCat. The CAI and GC contents of the optimized nucleotide sequence of the vaccine were 0.95 and 52.04%, respectively. Finally, in silico cloning of the vaccine construct into the pET-28 (+) vector was performed using the SnapGene software (Fig. 11).
In silico cloning of the multi-epitope vaccine into the pET28a (+) vector using SnapGene sofware free-trial (https://www.snapgene.com/free-trial/). The red section represents the vaccine construct and the black section shows the backbone of the vector.
Discussion
Cervical cancer, caused by HPV, is a public health crisis in both developing and developed countries25. Despite the availability of HPV prophylactic vaccines, developing a therapeutic vaccine for cervical cancer remains an essential need in public health26. Reverse vaccinology, an unconventional approach to the development of new vaccines that combines immunogenicity, immunogenicity, and bioinformatics, has attracted the attention of many researchers around the world27. This approach has been widely used to develop multi-epitope vaccines against a variety of organisms, including Helicobacter pylori28, Leishmania donovani29, Klebsiella pneumoniae30, hepatitis C virus31, Fasciola gigantica32, Tropheryma whipplei33, Elizabethkingia anopheles34, Candida auris35, dengue virus36, Zika virus37, and SARS-COV-238,39,40. In recent years, several studies have been published focusing on the design of multi-epitope vaccines for HPV. In the studies conducted by Negahdaripour et al., the HPV16 L2 protein was used to predict the epitope41,42, while Sarkar et al.43. and Mahmoudvand et al.44 used L1 protein to predict the epitope. In another study, Namvar et al. selected the E5 and E7 proteins from HPV16/18/31/45 as target proteins for epitope prediction45. The E6 and E7 proteins are key targets in the development of therapeutic vaccines against cervical cancer46 for a variety of reasons. First, HPV-associated malignancies and HPV-infected cells consistently express E6 and E7, whereas healthy cells do not14,15. Second, E6 and E7 proteins are essential for the initiation and maintenance of HPV-associated malignancies, cancer progression, and escape from the immune system47. Third, E6 and E7 proteins are viral antigens that are not subject to central tolerance by human immune systems48. Therefore, in the present study, the E6 and E7 oncoproteins from HPV16 were chosen as the target antigens for epitope prediction. The predicted epitopes were evaluated for antigenicity, toxicity, and allergenicity. In multi-epitope vaccines, the nature of the epitopes, adjuvants, and linkers, and their order and position in the chimeric sequence are all important. Since the physicochemical properties and the secondary and tertiary structures of the multi-epitope vaccine are determined by the construct’s structure, the nature of the epitopes, adjuvant, and linkers, and their arrangement and position in the multi-epitope vaccine are all important42. In this study, we used AAY linkers to fuse CTL epitopes, while we used GPGPG linkers to link HTL epitopes, similar to the studies by Khatoon et al.29, Tahir ul Qamar et al.49, and Tarang et al.50. GPGPG and AAY linkers promote epitope presentation, while they also reduce the formation of junctional epitopes51,52. The 50S ribosomal protein L7/L12 (Locus RL7_MYCTU) is a protein derived from Mycobacterium tuberculosis that has been shown in several studies to have an affinity for TLR453,54,55; hence, we used it as an adjuvant to improve the vaccine’s immunogenicity. The EAAAK linker decreases the connection with other protein areas, while increasing stability56,57.
The proposed vaccine construct was antigenic and non-allergic, indicating its effectiveness in eliciting robust immune responses without causing potentially-harmful allergic responses. The theoretical pI of the vaccine was found to be 8.33, indicating that the vaccine is basic in nature. The molecular weight of the vaccine was 32.01 kDa, which is appropriate since proteins with molecular weights less than 110 kDa are easier and quicker to purify58. The instability index of the vaccine was calculated to be 37.82, and as this value is below 40, the vaccine is considered a stable protein59. The half-life of our vaccine was determined to be 30 h in mammalian reticulocytes, while the half-life of the constructs designed in the study of Sarkar et al. is one hour43, indicating that our vaccine is exposed to the immune system for a longer period of time than the vaccines designed by Sarkar et al. The aliphatic index of the vaccine was calculated to be 76.43, which shows that it is thermostable60. The GRAVY value was -0.307, and a negative value for this parameter indicates that the vaccine is hydrophilic, it can interact with water molecules61. However, in the study of Negahdaripour et al.41. GRAVY was determined to be 0.252, and the use of micelles to increase vaccine interaction inside the polar environment of the body seems to be needed due to the vaccine's hydrophobic nature.
After building the vaccine’s three-dimensional structure, the refining process was used to improve its quality, getting it closer to the native structure. Model validation is necessary to compare the quality of the unrefined model with that of the refined model. The Ramachandran plot showed that 50.2% of the residues in the unrefined model were found in the favoured region, while 60.8% of the residues in the refined model were located in the favoured region, indicating the refined model’s improvement. The TLR4 immune receptor is expressed in human cervical cancer HeLa cells with a frequency 100 times higher than other TLRs, proving a correlation between TLR4 and cervical cancer progression62,63. Therefore, the molecular docking analysis of the vaccine was carried out with TLR4. The molecular docking results indicated that the vaccine interacted strongly with TLR4. The vaccine-TLR4 docked complex was also subjected to MD simulation to determine the vaccine construct’s stability. The RSMD plot of the proposed vaccine and TLR4 revealed that both were stable. According to the RMSF analysis, the vaccine construct had the lowest fluctuations in the regions with the most interactions with TLR4. Codon optimization was carried out in order to increase the expression of the vaccine candidate in E. coli (K12 strain). The vaccine sequence had a CAI value of 0.95 and a GC content of 52.04%. Since CAI values greater than 0.8 are considered to be good for expression in the target organism64, and since it is reported that a GC content between 30 and 70% is required for better expression61, the results of this section are satisfactory.
Materials and methods
Protein sequence retrieval
The reference sequence of E6 (NP_041325.1) and E7 (NP_041326.1) proteins from HPV16 were retrieved in FASTA format from NCBI database (https://www.ncbi.nlm.nih.gov/).
Identification and selection of T-cell epitopes
The NetCTL 1.2 server (http://www.cbs.dtu.dk/services/NetCTL/) was used to identify the CTL epitopes for the target proteins65. This server can predict CTL epitopes (9 mer), restricted to 12 MHC class I supertypes, including A1, A2, A3, A24, A26, B7, B8, B27, B39, B44, B58, and B62. A combination of three approaches proteasomal C-terminal cleavage, TAP transport efficiency, and MHC class-I binding affinity, is included in the prediction. TAP transport efficiency is evaluated using a weight matrix, while MHC-I binding and proteasoma C-terminal cleavage are predicted using artificial neural networks. In this study, the threshold value for epitope prediction was set at 0.75.
The NetMHCII 2.3 server (http://www.cbs.dtu.dk/services/NetMHCII/) was used to identify the HTL epitopes66. This server predicts the binding of the HTL epitopes (15 mer) to HLA-DR, HLA-DQ, HLA-DP, and mouse MHC class II alleles using artificial neural networks. In this study, the thresholds for strong and weak binders were set at 2% and 10%, respectively.
Due to the large number of epitopes, epitope screening for antigenicity, toxicity, and allergenicity is performed to select the best epitopes. The VaxiJen v2.0 server (http://www.ddg-pharmfac.net/vaxijen/VaxiJen/VaxiJen.html) was used to predict the antigenicity of the epitopes67,68,69. This server is capable of calculating the antigenicity of various microorganisms, such as bacteria, viruses, tumors, parasites, and fungi. The accuracy of the prediction by the VaxiJen v2.0 server is between 70 and 89%. In this analysis, the virus was selected as the target organism, and the antigenicity threshold was set at 0.4. Moreover, the ToxinPred server was used to predict epitope toxicity70. In this study, an SVM-based method (Swiss-Prot) was selected to predict toxicity. In addition, the AllerTOP v. 2.0 server (https://www.ddg-pharmfac.net/AllerTOP/method.html) was used to evaluate the allergenicity of the epitopes71. The strategy utilized in this server is based on the auto cross covariance (ACC) change of amino acid sequences into standard vectors of identical length72. It is important to note that not all HTL epitopes have the ability to induce the production of cytokines, and if produced, the cytokines produced by each may be different. Moreover, IL4pred (https://webs.iiitd.edu.in/raghava/il4pred/design.php) and IFNepitope (https://webs.iiitd.edu.in/raghava/ifnepitope/design.php) were used to predict IL-4 and IFN-γ inducing HTL epitopes, respectively. The SVM-based model and a threshold of 0.2 were selected to predict IL-4 inducing HTL epitopes73, and an SVM-based and IFN-gamma versus other cytokine models were selected to predict IFN-γ inducing HTL epitopes74.
Construction of the multi-epitope vaccine construct
Epitopes selected from the previous step were used to construct a multi-epitope vaccine. The HTL epitopes were linked using GPGPG linkers, whereas AAY linkers were used for the CTL epitopes. Linkers increase the representation and proper separation of the epitopes28. Moreover, glycine-rich linkers, such as GPGPG, also help improve solubility. Furthermore, the 50S ribosomal protein L7/L12 (Locus RL7_MYCTU) with accession no. P9WHE3 was selected as an adjuvant to enhance the immunogenicity of the vaccine candidate, and its amino acid sequence was attached by an EAAAK linker to the N-terminal of the chimeric sequences.
Evaluation of the antigenicity, allergenicity, and physicochemical properties of the vaccine
The assessment of antigenicity is an essential step in the process of designing vaccines. Two servers, VaxiJen v2.0 and ANTIGENpro, were used to predict the antigenic behavior of the final vaccine construct. ANTIGENpro (http://scratch.proteomics.ics.uci.edu/) estimates protein antigenicity using five machine learning algorithms and multiple representations of the initial sequence75. In order to ensure that the vaccine was not allergenic, AllerTOP v. 2.0 was used to predict the allergenicity of the vaccine. In this study, we used the Expasy ProtParam server (https://web.expasy.org/protparam/) to predict various physicochemical parameters of the multi-epitope vaccine, including amino acid composition, theoretical pI, molecular weight, instability index, half-life, aliphatic index, and grand average of hydropathicity (GRAVY)59.
Prediction of the secondary structure
We used the Prabi server (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl?page=/NPSA/npsa_gor4.html) to predict the percentage of secondary structure elements in the vaccine construct. GOR IV is the prediction method used on this server, which has a mean accuracy of 64.4%76.
Tertiary structure modeling, refinement, and validation of the multi-epitope vaccine
The I-TASSER server (https://zhanglab.ccmb.med.umich.edu/I-TASSER/) was used to predict the 3D model of the multi-epitope vaccine. This server generates three-dimensional structures from the amino acid sequence by reassembling the excised parts from the threading templates, and it calculates the C-score to evaluate the accuracy of the predicted models22,77,78. The selected model was refined using the 3Drefine server (http://sysbio.rnet.missouri.edu/3Drefine/) to improve its structural quality. The algorithm used in the 3Drefine server includes a two-step process (1) the optimization of the hydrogen bonding network, and (2) the minimization of atomic energy by integrating physics into the force field79,80,81. Model validation was performed using the ProSA-web server (https://prosa.services.came.sbg.ac.at/prosa.php) and the SAVES v6.0 server (https://saves.mbi.ucla.edu/). The ProSA-web server computes the overall quality Z-score for the protein structure. If the Z-score is outside the characteristic range of the native proteins, it indicates that there may be errors in the protein structure82,83. The PROCHECK tool of the SAVES v6.0 server evaluates the stereochemical quality of a protein structure by checking the geometry of the residues and the overall structural geometry84,85.
Prediction of the B-cell epitopes
The most important elements in the immune system are B lymphocytes, which are responsible for antibody secretion, thus promoting long-term immunity 28. The prediction of linear B-cell epitopes was performed using the BCPREDS (B-cell epitope prediction server) (http://ailab-projects1.ist.psu.edu:8080/bcpred/predict.html). This server uses a subsequence kernel-based SVM classifier with an accuracy of 74.57% to predict linear B-cell epitopes86,87,88. Furthermore, the Ellipro server (http://tools.iedb.org/ellipro/) was used for the prediction of discontinuous B-cell epitopes. The ElliPro server uses residue clustering algorithms along with the Tornton’s method for predicting discontinuous B-cell epitopes. The server assigns a score to each of the predicted epitopes, which is defined as a PI (protrusion index) value89.
Molecular docking
Molecular docking is one of the computational methods used to evaluate the interaction between two molecules and find the best orientation of a ligand in a complex. The ClusPro 2.0 server (https://cluspro.org/login.php) was used to evaluate the interaction between the vaccine construct and TLR4 (PDB ID: 4G8A)90,91,92,93. The refined model of the vaccine construct, as the ligand, and TLR4, as the receptor, were submitted to the server. The LigPlot program was used to illustrate the bonds that formed between the residues of the vaccine construct and TLR4 in the docked complex24.
MD simulation
The MD simulation was performed using the GROMACS 2019.6 software. Using Newton’s laws of atomic and molecular motion, the software predicts the behavior of ligands and receptors over a specific period of time94,95,96. The ff99SB force field was used to prepare the input structure. The surface charge of the structure was then neutralized using sodium and chloride ions. The gmx solvate software was also used to insert the protein into a layer of TIP3P water molecules with a thickness of 10 angstroms. To eliminate van der Waals interactions and hydrogen bonds forming between the water and the complex molecules, the energy of the structures was minimized using the steepest descent method. Afterward, the system temperature was steadily increased from 0 to 300 K in a constant volume for 200 ps, and the system was then equilibrated at constant pressure. Finally, the root-mean-square deviation (RMSD) and root-mean-square fluctuation (RMSF) of the ligand and the receptor were calculated over a 40-ns timeframe.
Codon optimization and in silico cloning of the final vaccine construct
The Java Codon Adaptation Tool (JCat) (http://www.jcat.de/) was used for the back translation and codon optimization of the final vaccine construct97. The protein sequence of the vaccine was submitted to the JCat, and E. coli (K12 strain) was selected as the host organism to express the vaccine construct. This server calculates two parameters, the codon adaptive index (CAI) and the GC content, which are important for the evaluation of protein expression levels. The sequence of restriction sites for XhoI and BamHI restriction enzymes were introduced at the 5′ and 3′ ends of the vaccine construct, respectively, and the vaccine sequence was then cloned into the pET28 (+) vector using the SnapGene software (https://www.snapgene.com/free-trial/).
Conclusions
Cervical cancer, caused by HPV, has affected the health of millions of people worldwide. There is currently no effective therapeutic vaccine available to treat HPV infections. In this study, we have attempted to design a multi-epitope vaccine against cervical cancer using reverse vaccinology. CTL and HTL epitopes from the E6 and E7 proteins of HPV16 were identified Antigenicity, toxicity, and allergenicity of the predicted epitopes were assessed, and the best epitopes were merged using appropriate linkers and adjuvant. The designed vaccine was found to be both antigenic and non-allergenic, and its physicochemical properties were acceptable. Molecular docking was also performed to check the binding affinity of the vaccine construct with TLR-4 in the vaccine-TLR4 complex. The stability of the vaccine candidate was confirmed by MD simulation. At last, the expression and translation efficiency of the multi-epitope vaccine was evaluated. Although the findings of this study were very impressive, they must be validated in the wet lab and animal models.
References
Arbyn, M. et al. Estimates of incidence and mortality of cervical cancer in 2018: A worldwide analysis. Lancet Glob. Health 8, e191–e203 (2020).
De Sanjosé, S. et al. Worldwide prevalence and genotype distribution of cervical human papillomavirus DNA in women with normal cytology: A meta-analysis. Lancet. Infect. Dis 7, 453–459 (2007).
De Villiers, E.-M., Fauquet, C., Broker, T. R., Bernard, H.-U. & Zur Hausen, H. Classification of papillomaviruses. Virology 324, 17–27 (2004).
Halec, G. et al. Biological activity of probable/possible high-risk human papillomavirus types in cervical cancer. Int. J. Cancer 132, 63–71 (2013).
Bouvard, V. et al. A review of human carcinogens—Part B: Biological agents. Lancet Oncol. 10, 321–322 (2009).
de Villiers, E.-M. Heterogeneity of the human papillomavirus group. J. Virol. 63, 4898 (1989).
Mirabello, L. et al. The intersection of HPV epidemiology, genomics and mechanistic studies of HPV-mediated carcinogenesis. Viruses 10, 80 (2018).
Muñoz, N. et al. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348, 518–527 (2003).
Doorbar, J. et al. The biology and life-cycle of human papillomaviruses. Vaccine 30, F55–F70 (2012).
Egawa, N., Egawa, K., Griffin, H. & Doorbar, J. Human papillomaviruses; epithelial tropisms, and the development of neoplasia. Viruses 7, 3863–3890 (2015).
Munkhdelger, J. et al. Performance of HPV E6/E7 mRNA RT-qPCR for screening and diagnosis of cervical cancer with ThinPrep® Pap test samples. Exp. Mol. Pathol. 97, 279–284 (2014).
Narisawa-Saito, M. & Kiyono, T. Basic mechanisms of high-risk human papillomavirus-induced carcinogenesis: Roles of E6 and E7 proteins. Cancer Sci. 98, 1505–1511 (2007).
Chemes, L. B., Camporeale, G., Sánchez, I. E., de Prat-Gay, G. & Alonso, L. G. Cysteine-rich positions outside the structural zinc motif of human papillomavirus E7 provide conformational modulation and suggest functional redox roles. Biochemistry 53, 1680–1696 (2014).
Doorbar, J., Egawa, N., Griffin, H., Kranjec, C. & Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 25, 2–23 (2015).
Sanclemente, G. & Gill, D. Human papillomavirus molecular biology and pathogenesis. J. Eur. Acad. Dermatol. Venereol. 16, 231–240 (2002).
Wang, R. et al. Human papillomavirus vaccine against cervical cancer: Opportunity and challenge. Cancer Lett. 471, 88–102 (2020).
Hildesheim, A. et al. Effect of human papillomavirus 16/18 L1 viruslike particle vaccine among young women with preexisting infection: A randomized trial. JAMA 298, 743–753 (2007).
Rappuoli, R. Reverse vaccinology. Curr. Opin. Microbiol. 3, 445–450 (2000).
Suhrbier, A. Multi-epitope DNA vaccines. Immunol. Cell Biol. 75, 402–408 (1997).
Rueckert, C. & Guzmán, C. A. Vaccines: From empirical development to rational design. PLoS Pathog. 8, e1003001 (2012).
Nezafat, N., Eslami, M., Negahdaripour, M., Rahbar, M. R. & Ghasemi, Y. Designing an efficient multi-epitope oral vaccine against Helicobacter pylori using immunoinformatics and structural vaccinology approaches. Mol. BioSyst. 13, 699–713 (2017).
Roy, A., Kucukural, A. & Zhang, Y. I-TASSER: A unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738 (2010).
Pettersen, E. F. et al. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Wallace, A. C., Laskowski, R. A. & Thornton, J. M. LIGPLOT: A program to generate schematic diagrams of protein–ligand interactions. Protein Eng. Des. Sel. 8, 127–134 (1995).
Vu, M., Yu, J., Awolude, O. A. & Chuang, L. Cervical cancer worldwide. Curr. Probl. Cancer 42, 457–465 (2018).
Tsang, K. Y. et al. Identification and characterization of enhancer agonist human cytotoxic T-cell epitopes of the human papillomavirus type 16 (HPV16) E6/E7. Vaccine 35, 2605–2611 (2017).
Mora, M., Veggi, D., Santini, L., Pizza, M. & Rappuoli, R. Reverse vaccinology. Drug Discov. Today 8, 459–464 (2003).
Khan, M. et al. Immunoinformatics approaches to explore Helicobacter pylori proteome (Virulence Factors) to design B and T cell multi-epitope subunit vaccine. Sci. Rep. 9, 1–13 (2019).
Khatoon, N., Pandey, R. K. & Prajapati, V. K. Exploring Leishmania secretory proteins to design B and T cell multi-epitope subunit vaccine using immunoinformatics approach. Sci. Rep. 7, 1–12 (2017).
Dar, H. A. et al. Immunoinformatics-aided design and evaluation of a potential multi-epitope vaccine against Klebsiella pneumoniae. Vaccines 7, 88 (2019).
Khalid, H. & Ashfaq, U. A. Exploring HCV genome to construct multi-epitope based subunit vaccine to battle HCV infection: Immunoinformatics based approach. J. Biomed. Inform. 108, 103498 (2020).
Kalita, P., Lyngdoh, D. L., Padhi, A. K., Shukla, H. & Tripathi, T. Development of multi-epitope driven subunit vaccine against Fasciola gigantica using immunoinformatics approach. Int. J. Biol. Macromol. 138, 224–233 (2019).
Joshi, A. & Kaushik, V. In-silico proteomic exploratory quest: Crafting T-cell epitope vaccine against Whipple’s disease. Int. J. Pept. Res. Ther. 27, 169–179 (2021).
Nain, Z. et al. Proteome-wide screening for designing a multi-epitope vaccine against emerging pathogen Elizabethkingia anophelis using immunoinformatic approaches. J. Biomol. Struct. Dyn. 38, 4850–4867 (2020).
Akhtar, N., Joshi, A., Kaushik, V., Kumar, M. & Mannan, M.A.-U. In-silico design of a multivalent epitope-based vaccine against Candida auris. Microbial. Pathog. 155, 104879 (2021).
Krishnan, S., Joshi, A., Akhtar, N. & Kaushik, V. Immunoinformatics designed T cell multi epitope dengue peptide vaccine derived from non structural proteome. Microbial. Pathog. 150, 104728 (2021).
Shahid, F., Ashfaq, U. A., Javaid, A. & Khalid, H. Immunoinformatics guided rational design of a next generation multi epitope based peptide (MEBP) vaccine by exploring Zika virus proteome. Infect. Genet. Evol. 80, 104199 (2020).
Rahman, N. et al. Vaccine design from the ensemble of surface glycoprotein epitopes of SARS-CoV-2: An immunoinformatics approach. Vaccines 8, 423 (2020).
Singh, A., Thakur, M., Sharma, L. K. & Chandra, K. Designing a multi-epitope peptide based vaccine against SARS-CoV-2. Sci. Rep. 10, 1–12 (2020).
Safavi, A., Kefayat, A., Mahdevar, E., Abiri, A. & Ghahremani, F. Exploring the out of sight antigens of SARS-CoV-2 to design a candidate multi-epitope vaccine by utilizing immunoinformatics approaches. Vaccine 38, 7612–7628 (2020).
Negahdaripour, M. et al. A novel HPV prophylactic peptide vaccine, designed by immunoinformatics and structural vaccinology approaches. Infect. Genet. Evol. 54, 402–416 (2017).
Negahdaripour, M. et al. Structural vaccinology considerations for in silico designing of a multi-epitope vaccine. Infect. Genet. Evol. 58, 96–109 (2018).
Sarkar, B., Ullah, M. A. & Araf, Y. A systematic and reverse vaccinology approach to design novel subunit vaccines against Dengue virus type-1 (DENV-1) and human Papillomavirus-16 (HPV-16). Inform. Med. Unlocked 19, 100343 (2020).
Mahmoudvand, S. et al. In silico prediction of T-cell and B-cell epitopes of human papillomavirus type 16 L1 protein. Biotechnol. Appl. Biochem. 20, 20 (2021).
Namvar, A., Panahi, H. A., Agi, E. & Bolhassani, A. Development of HPV 16, 18, 31, 45 E5 and E7 peptides-based vaccines predicted by immunoinformatics tools. Biotech. Lett. 42, 403–418 (2020).
Yang, A., Farmer, E., Lin, J., Wu, T.-C. & Hung, C.-F. The current state of therapeutic and T cell-based vaccines against human papillomaviruses. Virus Res. 231, 148–165 (2017).
Yugawa, T. & Kiyono, T. Molecular mechanisms of cervical carcinogenesis by high-risk human papillomaviruses: Novel functions of E6 and E7 oncoproteins. Rev. Med. Virol. 19, 97–113 (2009).
Peng, S. et al. Development of DNA vaccine targeting E6 and E7 proteins of human papillomavirus 16 (HPV16) and HPV18 for immunotherapy in combination with recombinant vaccinia boost and PD-1 antibody. MBio 12, 20 (2021).
Tahir ul Qamar, M. et al. Multiepitope-based subunit vaccine design and evaluation against respiratory syncytial virus using reverse vaccinology approach. Vaccines 8, 288 (2020).
Tarang, S. et al. In silico design of a multivalent vaccine against Candida albicans. Sci. Rep. 10, 1–7 (2020).
Livingston, B. et al. A rational strategy to design multiepitope immunogens based on multiple Th lymphocyte epitopes. J. Immunol. 168, 5499–5506 (2002).
Nezafat, N. et al. Designing an efficient multi-epitope peptide vaccine against Vibrio cholerae via combined immunoinformatics and protein interaction based approaches. Comput. Biol. Chem. 62, 82–95 (2016).
Chatterjee, N., Ojha, R., Khatoon, N. & Prajapati, V. K. Scrutinizing Mycobacterium tuberculosis membrane and secretory proteins to formulate multiepitope subunit vaccine against pulmonary tuberculosis by utilizing immunoinformatic approaches. Int. J. Biol. Macromol. 118, 180–188 (2018).
Deng, H. et al. Development of a multivalent enterovirus subunit vaccine based on immunoinformatic design principles for the prevention of HFMD. Vaccine 38, 3671–3681 (2020).
Pandey, R. K. & Prajapati, V. K. Exploring sand fly salivary proteins to design multiepitope subunit vaccine to fight against visceral leishmaniasis. J. Cell. Biochem. 120, 1141–1155 (2019).
Arai, R., Ueda, H., Kitayama, A., Kamiya, N. & Nagamune, T. Design of the linkers which effectively separate domains of a bifunctional fusion protein. Protein Eng. 14, 529–532 (2001).
Pandey, R. K. et al. Exploring dual inhibitory role of febrifugine analogues against Plasmodium utilizing structure-based virtual screening and molecular dynamic simulation. J. Biomol. Struct. Dyn. 35, 791–804 (2017).
Barh, D. et al. Exoproteome and secretome derived broad spectrum novel drug and vaccine candidates in Vibrio cholerae targeted by Piper betel derived compounds. PLoS One 8, e52773 (2013).
Gasteiger, E. et al. Protein identification and analysis tools on the ExPASy server. Proteom. Protoc. Handb. 20, 571–607 (2005).
Ikai, A. Thermostability and aliphatic index of globular proteins. J. Biochem. 88, 1895 (1980).
Ali, M. et al. Exploring dengue genome to construct a multi-epitope based subunit vaccine by utilizing immunoinformatics approach to battle against dengue infection. Sci. Rep. 7, 1–13 (2017).
Wang, Y. et al. Expression and functional analysis of Toll-like receptor 4 in human cervical carcinoma. J. Membr. Biol. 247, 591–599 (2014).
Nishimura, M. & Naito, S. Tissue-specific mRNA expression profiles of human toll-like receptors and related genes. Biol. Pharm. Bull. 28, 886–892 (2005).
Morla, S., Makhija, A. & Kumar, S. Synonymous codon usage pattern in glycoprotein gene of rabies virus. Gene 584, 1–6 (2016).
Larsen, M. V. et al. Large-scale validation of methods for cytotoxic T-lymphocyte epitope prediction. BMC Bioinform. 8, 1–12 (2007).
Jensen, K. K. et al. Improved methods for predicting peptide binding affinity to MHC class II molecules. Immunology 154, 394–406 (2018).
Doytchinova, I. A. & Flower, D. R. VaxiJen: A server for prediction of protective antigens, tumour antigens and subunit vaccines. BMC Bioinform. 8, 1–7 (2007).
Doytchinova, I. A. & Flower, D. R. Identifying candidate subunit vaccines using an alignment-independent method based on principal amino acid properties. Vaccine 25, 856–866 (2007).
Doytchinova, I. A. & Flower, D. R. Bioinformatic approach for identifying parasite and fungal candidate subunit vaccines. Open Vaccine J. 1, 4 (2008).
Gupta, S. et al. In silico approach for predicting toxicity of peptides and proteins. PLoS One 8, e73957 (2013).
Dimitrov, I., Bangov, I., Flower, D. R. & Doytchinova, I. AllerTOP v.2—a server for in silico prediction of allergens. J. Mol. Model. 20, 1–6 (2014).
Wold, S., Jonsson, J., Sjörström, M., Sandberg, M. & Rännar, S. DNA and peptide sequences and chemical processes multivariately modelled by principal component analysis and partial least-squares projections to latent structures. Anal. Chim. Acta 277, 239–253 (1993).
Dhanda, S. K., Gupta, S., Vir, P. & Raghava, G. Prediction of IL4 inducing peptides. Clin. Dev. Immunol. 2013, 20 (2013).
Dhanda, S. K., Vir, P. & Raghava, G. P. Designing of interferon-gamma inducing MHC class-II binders. Biol. Direct 8, 1–15 (2013).
Magnan, C. N. et al. High-throughput prediction of protein antigenicity using protein microarray data. Bioinformatics 26, 2936–2943 (2010).
Garnier, J. GOR secondary structure prediction method version IV. Methods Enzym. RF Doolittle Ed. 266, 540–553 (1998).
Yang, J. et al. The I-TASSER suite: Protein structure and function prediction. Nat. Methods 12, 7–8 (2015).
Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 9, 1–8 (2008).
Bhattacharya, D. & Cheng, J. i3Drefine software for protein 3D structure refinement and its assessment in CASP10. PLoS One 8, e69648 (2013).
Bhattacharya, D., Nowotny, J., Cao, R. & Cheng, J. 3Drefine: An interactive web server for efficient protein structure refinement. Nucleic Acids Res. 44, W406–W409 (2016).
Bhattacharya, D. & Cheng, J. 3Drefine: Consistent protein structure refinement by optimizing hydrogen bonding network and atomic-level energy minimization. Proteins Struct. Funct. Bioinform. 81, 119–131 (2013).
Wiederstein, M. & Sippl, M. J. ProSA-web: Interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 35, W407–W410 (2007).
Sippl, M. J. Recognition of errors in three-dimensional structures of proteins. Proteins Struct. Funct. Bioinform. 17, 355–362 (1993).
Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. PROCHECK: A program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 26, 283–291 (1993).
Laskowski, R. A., Rullmann, J. A. C., MacArthur, M. W., Kaptein, R. & Thornton, J. M. AQUA and PROCHECK-NMR: Programs for checking the quality of protein structures solved by NMR. J. Biomol. NMR 8, 477–486 (1996).
Chen, J., Liu, H., Yang, J. & Chou, K.-C. Prediction of linear B-cell epitopes using amino acid pair antigenicity scale. Amino Acids 33, 423–428 (2007).
El-Manzalawy, Y., Dobbs, D. & Honavar, V. Predicting linear B-cell epitopes using string kernels. J. Mol. Recogn. Interdiscip. J. 21, 243–255 (2008).
El-Manzalawy, Y., Dobbs, D. & Honavar, V. Computational Systems Bioinformatics: Vol 7 121–132 (World Scientific, 2008).
Ponomarenko, J. et al. ElliPro: A new structure-based tool for the prediction of antibody epitopes. BMC Bioinform. 9, 1–8 (2008).
Desta, I. T., Porter, K. A., Xia, B., Kozakov, D. & Vajda, S. Performance and its limits in rigid body protein-protein docking. Structure 28, 1071-1081.e1073 (2020).
Kozakov, D. et al. The ClusPro web server for protein–protein docking. Nat. Protoc. 12, 255 (2017).
Vajda, S. et al. New additions to the Clus Pro server motivated by CAPRI. Proteins Struct. Funct. Bioinform. 85, 435–444 (2017).
Kozakov, D. et al. How good is automated protein docking?. Proteins Struct. Funct. Bioinform. 81, 2159–2166 (2013).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1, 19–25 (2015).
Páll, S., Abraham, M. J., Kutzner, C., Hess, B. & Lindahl, E. International Conference on Exascale Applications and Software 3–27 (Springer, 2014).
Pronk, S. et al. GROMACS 4.5: A high-throughput and highly parallel open source molecular simulation toolkit. Bioinformatics 29, 845–854 (2013).
Grote, A. et al. JCat: A novel tool to adapt codon usage of a target gene to its potential expression host. Nucleic Acids Res. 33, W526–W531 (2005).
Acknowledgements
The authors would like to acknowledge the Research Deputy of Shahrekord University of Medical Sciences for providing facilities for this study.
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sanami, S., Azadegan-Dehkordi, F., Rafieian-Kopaei, M. et al. Design of a multi-epitope vaccine against cervical cancer using immunoinformatics approaches. Sci Rep 11, 12397 (2021). https://doi.org/10.1038/s41598-021-91997-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-91997-4
This article is cited by
-
Designing a T-cell epitope-based vaccine using in silico approaches against the Sal k 1 allergen of Salsola kali plant
Scientific Reports (2024)
-
Immunoinformatics-aided design of a new multi-epitope vaccine adjuvanted with domain 4 of pneumolysin against Streptococcus pneumoniae strains
BMC Bioinformatics (2023)
-
Predicting Plasmodium falciparum kinase inhibitors from antimalarial medicinal herbs using computational modeling approach
In Silico Pharmacology (2023)
-
Designing a novel SOX9 based multi-epitope vaccine to combat metastatic triple-negative breast cancer using immunoinformatics approach
Molecular Diversity (2023)
-
Design of a potential Sema4A-based multi-epitope vaccine to combat triple-negative breast cancer: an immunoinformatic approach
Medical Oncology (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.