Abstract
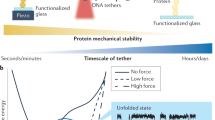
The reversible unfolding and refolding of proteins is a regulatory mechanism of tissue elasticity and signalling used by cells to sense and adapt to extracellular and intracellular mechanical forces. However, most of these proteins exhibit low mechanical stability, posing technical challenges to the characterization of their conformational dynamics under force. Here, we detail step-by-step instructions for conducting single-protein nanomechanical experiments using ultra-stable magnetic tweezers, which enable the measurement of the equilibrium conformational dynamics of single proteins under physiologically relevant low forces applied over biologically relevant timescales. We report the basic principles determining the functioning of the magnetic tweezer instrument, review the protein design strategy and the fluid chamber preparation and detail the procedure to acquire and analyze the unfolding and refolding trajectories of individual proteins under force. This technique adds to the toolbox of single-molecule nanomechanical techniques and will be of particular interest to those interested in proteins involved in mechanosensing and mechanotransduction. The procedure takes 4 d to complete, plus an additional 6 d for protein cloning and production, requiring basic expertise in molecular biology, surface chemistry and data analysis.
Key points
-
Ultra-stable magnetic tweezers are used for measuring the conformational dynamics of individual proteins at physiologically relevant low forces and over long timescales.
-
Magnetic fields are created by using either permanent magnets or a tape head, which generates precisely calibrated forces for pulling single proteins tethered between a superparamagnetic bead and a functionalized glass substrate.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout










Similar content being viewed by others
Data availability
Example data from Figs. 7 and 10 can be found as Supplementary Data. Modified pFN18a plasmids from Fig. 5 are available in Addgene (pFN18A-HaloTag-Biotin: Addgene plasmid #206039; pFN18A-HaloTag-SpyCatcher Addgene plasmid #206041). Other data that support the plots within this paper are available from the corresponding author upon reasonable request.
Code availability
Scripts for the fluctuation analysis are included in the Supplementary Data. The data acquisition code can be accessed at https://doi.org/10.5281/zenodo.8092186.
References
Swift, J. et al. Nuclear lamin-A scales with tissue stiffness and enhances matrix-directed differentiation. Science 341, 1240104 (2013).
De Belly, H., Paluch, E. K. & Chalut, K. J. Interplay between mechanics and signalling in regulating cell fate. Nat. Rev. Mol. Cell Biol. 23, 465–480 (2022).
Iskratsch, T., Wolfenson, H. & Sheetz, M. P. Appreciating force and shape—the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 15, 825–833 (2014).
Paul, C. D., Mistriotis, P. & Konstantopoulos, K. Cancer cell motility: lessons from migration in confined spaces. Nat. Rev. Cancer 17, 131–140 (2017).
Dumortier, J. G. et al. Hydraulic fracturing and active coarsening position the lumen of the mouse blastocyst. Science 365, 465–468 (2019).
Mongera, A. et al. A fluid-to-solid jamming transition underlies vertebrate body axis elongation. Nature 561, 401–405 (2018).
Pesce, M. et al. Cardiac fibroblasts and mechanosensation in heart development, health and disease. Nat. Rev. Cardiol. 20, 309–324 (2022).
Kefauver, J. M., Ward, A. B. & Patapoutian, A. Discoveries in structure and physiology of mechanically activated ion channels. Nature 587, 567–576 (2020).
Huse, M. Mechanical forces in the immune system. Nat. Rev. Immunol. 17, 679–690 (2017).
Romani, P., Valcarcel-Jimenez, L., Frezza, C. & Dupont, S. Crosstalk between mechanotransduction and metabolism. Nat. Rev. Mol. Cell Biol. 22, 22–38 (2021).
Romani, P. et al. Mitochondrial fission links ECM mechanotransduction to metabolic redox homeostasis and metastatic chemotherapy resistance. Nat. Cell Biol. 24, 168–180 (2022).
Jaalouk, D. E. & Lammerding, J. Mechanotransduction gone awry. Nat. Rev. Mol. Cell Biol. 10, 63–73 (2009).
Chemla, Y. R. et al. Mechanism of force generation of a viral DNA packaging motor. Cell 122, 683–692 (2005).
Ranade, S. S., Syeda, R. & Patapoutian, A. Mechanically activated ion channels. Neuron 87, 1162–1179 (2015).
Neuman, K. C. & Nagy, A. Single-molecule force spectroscopy: optical tweezers, magnetic tweezers and atomic force microscopy. Nat. Methods 5, 491–505 (2008).
Krieg, M. et al. Atomic force microscopy-based mechanobiology. Nat. Rev. Phys. 1, 41–57 (2018).
Bustamante, C. J., Chemla, Y. R., Liu, S. & Wang, M. D. Optical tweezers in single-molecule biophysics. Nat. Rev. Methods Prim. 1, 25 (2021).
Popa, I., Kosuri, P., Alegre-Cebollada, J., Garcia-Manyes, S. & Fernandez, J. M. Force dependency of biochemical reactions measured by single-molecule force-clamp spectroscopy. Nat. Protoc. 8, 1261–1276 (2013).
Li, H. et al. Reverse engineering of the giant muscle protein titin. Nature 418, 998–1002 (2002).
Echelman, D. J., Lee, A. Q. & Fernandez, J. M. Mechanical forces regulate the reactivity of a thioester bond in a bacterial adhesin. J. Biol. Chem. 292, 8988–8997 (2017).
Milles, L. F., Schulten, K., Gaub, H. E. & Bernardi, R. C. Molecular mechanism of extreme mechanostability in a pathogen adhesin. Science 359, 1527–1533 (2018).
Mora, M., Stannard, A. & Garcia-Manyes, S. The nanomechanics of individual proteins. Chem. Soc. Rev. 49, 6816–6832 (2020).
Dudko, O. K., Hummer, G. & Szabo, A. Theory, analysis, and interpretation of single-molecule force spectroscopy experiments. Proc. Natl Acad. Sci. USA 105, 15755–15760 (2008).
Cecconi, C., Shank, E. A., Bustamante, C. & Marqusee, S. Direct observation of the three-state folding of a single protein molecule. Science 309, 2057–2060 (2005).
Stigler, J., Ziegler, F., Gieseke, A., Gebhardt, J. C. & Rief, M. The complex folding network of single calmodulin molecules. Science 334, 512–516 (2011).
Neupane, K., Manuel, A. P. & Woodside, M. T. Protein folding trajectories can be described quantitatively by one-dimensional diffusion over measured energy landscapes. Nat. Phys. 12, 700–703 (2016).
Woodside, M. T. & Block, S. M. Reconstructing folding energy landscapes by single-molecule force spectroscopy. Annu. Rev. Biophys. 43, 19–39 (2014).
Kaiser, C. M., Goldman, D. H., Chodera, J. D., Tinoco, I. Jr. & Bustamante, C. The ribosome modulates nascent protein folding. Science 334, 1723–1727 (2011).
Lipfert, J., Kerssemakers, J. W., Jager, T. & Dekker, N. H. Magnetic torque tweezers: measuring torsional stiffness in DNA and RecA-DNA filaments. Nat. Methods 7, 977–980 (2010).
Ding, F. et al. Single-molecule mechanical identification and sequencing. Nat. Methods 9, 367–372 (2012).
Hodeib, S. et al. Single molecule studies of helicases with magnetic tweezers. Methods 105, 3–15 (2016).
Lionnet, T. et al. Magnetic trap construction. Cold Spring Harb. Protoc. 2012, 133–138 (2012).
Rivas-Pardo, J. A. et al. Work done by titin protein folding assists muscle contraction. Cell Rep. 14, 1339–1347 (2016).
Chen, H. et al. Dynamics of equilibrium folding and unfolding transitions of titin immunoglobulin domain under constant forces. J. Am. Chem. Soc. 137, 3540–3546 (2015).
Bauer, M. S. et al. A tethered ligand assay to probe SARS-CoV-2:ACE2 interactions. Proc. Natl Acad. Sci. USA 119, e2114397119 (2022).
Yao, M. et al. The mechanical response of talin. Nat. Commun. 7, 11966 (2016).
Popa, I. et al. A HaloTag anchored ruler for week-long studies of protein dynamics. J. Am. Chem. Soc. 138, 10546–10553 (2016).
Lof, A. et al. Multiplexed protein force spectroscopy reveals equilibrium protein folding dynamics and the low-force response of von Willebrand factor. Proc. Natl Acad. Sci. USA 116, 18798–18807 (2019).
Zhao, X., Zeng, X., Lu, C. & Yan, J. Studying the mechanical responses of proteins using magnetic tweezers. Nanotechnology 28, 414002 (2017).
Choi, H. K. et al. Watching helical membrane proteins fold reveals a common N-to-C-terminal folding pathway. Science 366, 1150–1156 (2019).
Choi, H. K. et al. Evolutionary balance between foldability and functionality of a glucose transporter. Nat. Chem. Biol. 18, 713–723 (2022).
Choi, H. K., Kim, H. G., Shon, M. J. & Yoon, T. Y. High-resolution single-molecule magnetic tweezers. Annu. Rev. Biochem. 91, 33–59 (2022).
Smith, S. B., Finzi, L. & Bustamante, C. Direct mechanical measurements of the elasticity of single DNA molecules by using magnetic beads. Science 258, 1122–1126 (1992).
Strick, T. R., Allemand, J. F., Bensimon, D., Bensimon, A. & Croquette, V. The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837 (1996).
Gosse, C. & Croquette, V. Magnetic tweezers: micromanipulation and force measurement at the molecular level. Biophys. J. 82, 3314–3329 (2002).
Maier, B., Bensimon, D. & Croquette, V. Replication by a single DNA polymerase of a stretched single-stranded DNA. Proc. Natl Acad. Sci. USA 97, 12002–12007 (2000).
Dekker, N. H. et al. The mechanism of type IA topoisomerases. Proc. Natl Acad. Sci. USA 99, 12126–12131 (2002).
Crut, A., Koster, D. A., Seidel, R., Wiggins, C. H. & Dekker, N. H. Fast dynamics of supercoiled DNA revealed by single-molecule experiments. Proc. Natl Acad. Sci. USA 104, 11957–11962 (2007).
England, C. G., Luo, H. & Cai, W. HaloTag technology: a versatile platform for biomedical applications. Bioconjug. Chem. 26, 975–986 (2015).
Zakeri, B. et al. Peptide tag forming a rapid covalent bond to a protein, through engineering a bacterial adhesin. Proc. Natl Acad. Sci. USA 109, E690–E697 (2012).
Tapia-Rojo, R., Eckels, E. C. & Fernandez, J. M. Ephemeral states in protein folding under force captured with a magnetic tweezers design. Proc. Natl Acad. Sci. USA 116, 7873–7878 (2019).
Stannard, A. et al. Molecular fluctuations as a ruler of force-induced protein conformations. Nano Lett. 21, 2953–2961 (2021).
Tapia-Rojo, R., Alonso-Caballero, A. & Fernandez, J. M. Direct observation of a coil-to-helix contraction triggered by vinculin binding to talin. Sci. Adv. 6, eaaz4707 (2020).
Franz, F. et al. Allosteric activation of vinculin by talin. Nat. Commun. 14, 4311 (2023).
Tapia-Rojo, R. et al. Enhanced statistical sampling reveals microscopic complexity in the talin mechanosensor folding energy landscape. Nat. Phys. 19, 52–60 (2023).
Austen, K. et al. Extracellular rigidity sensing by talin isoform-specific mechanical linkages. Nat. Cell Biol. 17, 1597–1606 (2015).
Riveline, D. et al. Focal contacts as mechanosensors: externally applied local mechanical force induces growth of focal contacts by an mDia1-dependent and ROCK-independent mechanism. J. Cell Biol. 153, 1175–1186 (2001).
Grashoff, C. et al. Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263–266 (2010).
Garcia-Manyes, S. et al. Single-molecule force spectroscopy predicts a misfolded, domain-swapped conformation in human γD-crystallin protein. J. Biol. Chem. 291, 4226–4235 (2016).
Mora, M. et al. A single-molecule strategy to capture non-native intramolecular and intermolecular protein disulfide bridges. Nano Lett. 22, 3922–3930 (2022).
Petrosyan, R., Patra, S., Rezajooei, N., Garen, C. R. & Woodside, M. T. Unfolded and intermediate states of PrP play a key role in the mechanism of action of an antiprion chaperone. Proc. Natl Acad. Sci. USA 118, e2010213118 (2021).
Gupta, A. N. et al. Pharmacological chaperone reshapes the energy landscape for folding and aggregation of the prion protein. Nat. Commun. 7, 12058 (2016).
Sen Mojumdar, S. et al. Partially native intermediates mediate misfolding of SOD1 in single-molecule folding trajectories. Nat. Commun. 8, 1881 (2017).
Yao, M. et al. Force-dependent conformational switch of alpha-catenin controls vinculin binding. Nat. Commun. 5, 4525 (2014).
Dahal, N., Sharma, S., Phan, B., Eis, A. & Popa, I. Mechanical regulation of talin through binding and history-dependent unfolding. Sci. Adv. 8, eabl7719 (2022).
Kemmerich, F. E. et al. Simultaneous single-molecule force and fluorescence sampling of DNA nanostructure conformations using magnetic tweezers. Nano Lett. 16, 381–386 (2016).
Ivanov, I. E. et al. Multimodal measurements of single-molecule dynamics using FluoRBT. Biophys. J. 114, 278–282 (2018).
Tapia-Rojo, R., Alonso-Caballero, A. & Fernandez, J. M. Talin folding as the tuning fork of cellular mechanotransduction. Proc. Natl Acad. Sci. USA 117, 21346–21353 (2020).
Alonso-Caballero, A. et al. Protein folding modulates the chemical reactivity of a Gram-positive adhesin. Nat. Chem. 13, 172–181 (2021).
Guo, H. A simple algorithm for fitting a gaussian function. In Streamlining Digital Signal Processing 297–305 (John Wiley & Sons, 2012).
Fonnum, G., Johansson, C., Molteberg, A., Mørup, S. & Aksnes, E. Characterisation of Dynabeads® by magnetization measurements and Mössbauer spectroscopy. J. Magn. Magn. Mater. 293, 41–47 (2005).
Ostrofet, E., Papini, F. S. & Dulin, D. Correction-free force calibration for magnetic tweezers experiments. Sci. Rep. 8, 15920 (2018).
Buschow, K. H. J., Long, G. J. & Grandjean, F. High Density Digital Recording (Springer, 1993).
Liu, R., Garcia-Manyes, S., Sarkar, A., Badilla, C. L. & Fernandez, J. M. Mechanical characterization of protein L in the low-force regime by electromagnetic tweezers/evanescent nanometry. Biophys. J. 96, 3810–3821 (2009).
Valle-Orero, J. et al. Proteins breaking bad: a free energy perspective. J. Phys. Chem. Lett. 8, 3642–3647 (2017).
Alegre-Cebollada, J., Badilla, C. L. & Fernandez, J. M. Isopeptide bonds block the mechanical extension of pili in pathogenic Streptococcus pyogenes. J. Biol. Chem. 285, 11235–11242 (2010).
Schlierf, M., Li, H. & Fernandez, J. M. The unfolding kinetics of ubiquitin captured with single-molecule force-clamp techniques. Proc. Natl Acad. Sci. USA 101, 7299–7304 (2004).
Evans, E. & Ritchie, K. Dynamic strength of molecular adhesion bonds. Biophys. J. 72, 1541–1555 (1997).
Zhang, Y., Jiao, J. & Rebane, A. A. Hidden Markov modeling with detailed balance and its application to single protein folding. Biophys. J. 111, 2110–2124 (2016).
McKinney, S. A., Joo, C. & Ha, T. Analysis of single-molecule FRET trajectories using hidden Markov modeling. Biophys. J. 91, 1941–1951 (2006).
Dudko, O. K., Hummer, G. & Szabo, A. Intrinsic rates and activation free energies from single-molecule pulling experiments. Phys. Rev. Lett. 96, 108101 (2006).
Bullerjahn, J. T., Sturm, S. & Kroy, K. Theory of rapid force spectroscopy. Nat. Commun. 5, 4463 (2014).
Acknowledgements
We are deeply grateful to J. Fernandez and C. Badilla (Columbia University) for their pioneering work on technique development and protein engineering and for their legacy in the field. We thank S. Board, J. Walker and P. Paracuellos for help in protein expression and purification. This work was supported in part by the Francis Crick Institute, which receives its core funding from Cancer Research U.K. (CC0102), the U.K. Medical Research Council (CC0102) and the Wellcome Trust (CC0102). R.T.-R. is the recipient of a King’s Prize Fellowship. This work was supported by the European Commission (Mechanocontrol, Grant Agreement 731957), BBSRC sLoLa (BB/V003518/1), Leverhulme Trust Research Leadership Award RL 2016-015, Wellcome Trust Investigator Award 212218/Z/18/Z and Royal Society Wolfson Fellowship RSWF/R3/183006 to S.G.-M.
Author information
Authors and Affiliations
Contributions
R.T.-R, M.M. and S.G.-M wrote the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks Tony Huang and the other, anonymous, reviewer(s) for their contribution to the peer review process of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Tapia-Rojo, R. et al. Nat. Phys. 19, 52–60 (2023): https://doi.org/10.1038/s41567-022-01808-4
Extended data
Extended Data Fig. 1 Calculating the stiffness of the magnetic trap.
Stiffness of the magnetic trap created by the N52 magnets (voice-coil configuration) (a) and magnetic tape head (b). The magnetic trap stiffnesses can be simply calculated as dF/dz, where z is the distance between the gap (magnets or tape head) and the magnetic bead. Because of the nonlinearity of F(z), the stiffness changes over the control parameter (magnet position or electric current), but in the operating regime of the trap this results in a very soft trap (~10−4 pN/nm), resulting in effective force clamp conditions (no appreciable change in force over the range in which the bead moves).
Extended Data Fig. 2 Calibration of the tweezers.
Calibration of the voice coil-based (a) or tape head–based (b) magnetic tweezers using the worm-like chain model for polymer elasticity (left) and comparison of the calibration using the worm-like chain (WLC) and freely jointed chain (FJC) (right). The FJC gives a lower contour length (ΔLc = 16.3 nm) compared to the WLC (ΔLc = 18.6 nm). All error bars are s.d.
Extended Data Fig. 3 Tape head and magnets.
The magnetic tape head and voice-coil-mounted permanent magnets with a magnification of the gap region.
Supplementary information
Supplementary Data 1
Raw traces from talin R3IVVI pulled at 1 pN/s and protein L pulled at 5 and 10 pN/s
Supplementary Data 2
Raw trace and fluctuation analysis of talin R3IVVI pulled at 8.5 pN
Supplementary Videos 1–3
1, how to pull on a protein by using single-molecule magnetic tweezers; 2, how to assemble the fluid chambers; 3, how to calibrate the distance between the bottom glass cover slide and the magnets
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tapia-Rojo, R., Mora, M. & Garcia-Manyes, S. Single-molecule magnetic tweezers to probe the equilibrium dynamics of individual proteins at physiologically relevant forces and timescales. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-00965-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-024-00965-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.