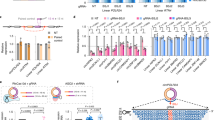
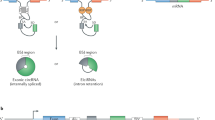
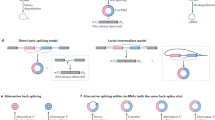
Abstract
Circular RNAs (circRNAs) are covalently enclosed, single-stranded RNAs produced by back-splicing of pre-mRNA exons that have recently emerged as an important class of molecules in gene expression regulation. circRNAs share overlapping sequences with their cognate linear mRNAs except the back-splicing junction (BSJ) sites. This feature makes it difficult to discriminate between the functions of circRNAs and their cognate mRNAs. We previously reported that the programmable RNA-guided, RNA-targeting CRISPR–Cas13 (RfxCas13d) system effectively and specifically discriminates circRNAs from mRNAs by using guide RNAs (gRNAs) targeting sequences across BSJ sites. Here, we describe a detailed protocol based on this RfxCas13d/BSJ-gRNA system for large-scale functional circRNA screening in human cell lines. The protocol includes gRNA library design, construction and transduction, analysis of screening results and validation of functional circRNA candidates. In total, it takes ~3–4 months of collaborative work between a well-trained molecular biologist and a bioinformatic expert. This protocol can be applied both in cells and in vivo to identify highly expressed circRNAs affecting cell growth, either in unperturbed conditions or under environmental stimulation, without disturbing their cognate linear mRNAs.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
The data used to generate the example results shown in Fig. 5 were originally published in ref. 23. All sequencing datasets have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GSE149690, GSE149691 and GSE149692) and National Omics Data Encyclopedia (OEP000887, OEP000888 and OEP000889). All the plasmids, cell lines and reagents presented in the paper are available on request from the corresponding author.
Code availability
The custom Perl and Shell scripts for the computational pipeline of the CDCscreen in this paper are available at https://github.com/YangLab/CDCscreen.
References
Hsu, M. T. & Coca-Prados, M. Electron microscopic evidence for the circular form of RNA in the cytoplasm of eukaryotic cells. Nature 280, 339–340 (1979).
Cocquerelle, C., Mascrez, B., Hetuin, D. & Bailleul, B. Mis-splicing yields circular RNA molecules. FASEB J. 7, 155–160 (1993).
Capel, B. et al. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell 73, 1019–1030 (1993).
Cocquerelle, C., Daubersies, P., Majerus, M. A., Kerckaert, J. P. & Bailleul, B. Splicing with inverted order of exons occurs proximal to large introns. EMBO J. 11, 1095–1098 (1992).
Nigro, J. M. et al. Scrambled exons. Cell 64, 607–613 (1991).
Yang, L., Duff, M. O., Graveley, B. R., Carmichael, G. G. & Chen, L. L. Genomewide characterization of non-polyadenylated RNAs. Genome Biol. 12, R16 (2011).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Jeck, W. R. et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA 19, 141–157 (2013).
Zhang, X. O. et al. Complementary sequence-mediated exon circularization. Cell 159, 134–147 (2014).
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N. & Brown, P. O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PloS One 7, e30733 (2012).
Westholm, J. O. et al. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9, 1966–1980 (2014).
Guo, J. U., Agarwal, V., Guo, H. & Bartel, D. P. Expanded identification and characterization of mammalian circular RNAs. Genome Biol. 15, 409 (2014).
Ye, C. Y., Chen, L., Liu, C., Zhu, Q. H. & Fan, L. Widespread noncoding circular RNAs in plants. N. Phytol. 208, 88–95 (2015).
Ivanov, A. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10, 170–177 (2015).
Fan, X. et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148 (2015).
Huang, R. et al. Circular RNA HIPK2 regulates astrocyte activation via cooperation of autophagy and ER stress by targeting MIR124-2HG. Autophagy 13, 1722–1741 (2017).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Piwecka, M. et al. Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 (2017).
Li, Q. et al. CircACC1 regulates assembly and activation of AMPK complex under metabolic stress. Cell Metab. 30, 157–173.e7 (2019).
Du, W. W. et al. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 44, 2846–2858 (2016).
Liu, Y. L. et al. Back-spliced RNA from retrotransposon binds to centromere and regulates centromeric chromatin loops in maize. PLoS Biol. 18, e3000582 (2020).
Conn, V. M. et al. A circRNA from SEPALLATA3 regulates splicing of its cognate mRNA through R-loop formation. Nat. Plants 3, 17053 (2017).
Li, S. Q. et al. Screening for functional circular RNAs using the CRISPR-Cas13 system. Nat. Methods 18, 51–59 (2021).
Abdelmohsen, K. et al. Identification of HuR target circular RNAs uncovers suppression of PABPN1 translation by CircPABPN1. RNA Biol. 14, 361–369 (2017).
Liu, C. X. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880.e21 (2019).
Li, X. et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell 67, 214–227.e7 (2017).
Xia, P. et al. A circular RNA protects dormant hematopoietic stem cells from DNA sensor cGAS-mediated exhaustion. Immunity 48, 688–701.e7 (2018).
Legnini, I. et al. Circ-ZNF609 is a circular RNA that can be translated and functions in myogenesis. Mol. Cell 66, 22–37.e9 (2017).
Li, X., Yang, L. & Chen, L. L. The biogenesis, functions, and challenges of circular RNAs. Mol. Cell 71, 428–442 (2018).
Zhang, Y. et al. The biogenesis of nascent circular RNAs. Cell Rep. 15, 611–624 (2016).
Kushawah, G. et al. CRISPR-Cas13d induces efficient mRNA knockdown in animal embryos. Dev. Cell 54, 805–817.e7 (2020).
Konermann, S. et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676.e14 (2018).
Zhang, Y. et al. Optimized RNA-targeting CRISPR/Cas13d technology outperforms shRNA in identifying functional circRNAs. Genome Biol. 22, 41 (2021).
Ai, Y., Liang, D. & Wilusz, J. E. CRISPR/Cas13 effectors have differing extents of off-target effects that limit their utility in eukaryotic cells. Nucleic Acids Res. In press. https://doi.org/10.1093/nar/gkac159 (2022).
Shmakov, S. et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol. Cell 60, 385–397 (2015).
East-Seletsky, A. et al. Two distinct RNase activities of CRISPR-C2c2 enable guide-RNA processing and RNA detection. Nature 538, 270–273 (2016).
Abudayyeh, O. O. et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science 353, aaf5573 (2016).
Kannan, S. et al. Compact RNA editors with small Cas13 proteins. Nat. Biotechnol. 40, 194–197 (2022).
Cox, D. B. T. et al. RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017).
Abudayyeh, O. O. et al. RNA targeting with CRISPR-Cas13. Nature 550, 280–284 (2017).
Myhrvold, C. et al. Field-deployable viral diagnostics using CRISPR-Cas13. Science 360, 444–448 (2018).
Gootenberg, J. S. et al. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science 360, 439–444 (2018).
Zhang, C. et al. Structural basis for the RNA-guided ribonuclease activity of CRISPR-Cas13d. Cell 175, 212–223.e17 (2018).
Wessels, H. H. et al. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 38, 722–727 (2020).
Cong, L. et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013).
Garneau, J. E. et al. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature 468, 67––71 (2010).
Ma, X. K. et al. CIRCexplorer3: a CLEAR pipeline for direct comparison of circular and linear RNA expression. Genomics Proteom. Bioinforma. 17, 511–521 (2019).
Joung, J. et al. Genome-scale CRISPR-Cas9 knockout and transcriptional activation screening. Nat. Protoc. 12, 828–863 (2017).
Li, W. et al. MAGeCK enables robust identification of essential genes from genome-scale CRISPR/Cas9 knockout screens. Genome Biol. 15, 554 (2014).
Shalem, O. et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science 343, 84–87 (2014).
Ye, L. et al. In vivo CRISPR screening in CD8 T cells with AAV-Sleeping Beauty hybrid vectors identifies membrane targets for improving immunotherapy for glioblastoma. Nat. Biotechnol. 37, 1302–1313 (2019).
Mosmann, T. Rapid colorimetric assay for cellular growth and survival: application to proliferation and cytotoxicity assays. J. Immunol. Methods 65, 55–63 (1983).
Bernabei, P. A. et al. In vitro chemosensitivity testing of leukemic cells: development of a semiautomated colorimetric assay. Hematol. Oncol. 7, 243–253 (1989).
Acknowledgements
We thank W. Xue for providing sequencing data-analysis support. This study was supported by the National Key R&D Program of China (2021YFA1300501), the National Natural Science Foundation of China (NSFC) (31725009, 91940303 and 31821004), the Center for Excellence in Molecular Cell Science (CEMCS) (2020DF03) and the HHMI International Program (55008728) to L.-L.C.; and NSFC (32101042) to S.L. L.-L.C. acknowledges support from the Xplorer Prize.
Author information
Authors and Affiliations
Contributions
L.-L.C. supervised and conceived the project. L.-L.C., S.L. and H.W. designed the experiments. S.L. and H.W. performed the experiments. L.-L.C., S.L. and H.W. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Protocols thanks David Gilot and Jun Wei Pek for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Li, S. et al. Nat. Methods 18, 51–59 (2021): https://doi.org/10.1038/s41592-020-01011-4
Rights and permissions
About this article
Cite this article
Li, S., Wu, H. & Chen, LL. Screening circular RNAs with functional potential using the RfxCas13d/BSJ-gRNA system. Nat Protoc 17, 2085–2107 (2022). https://doi.org/10.1038/s41596-022-00715-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-022-00715-5
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.