Abstract
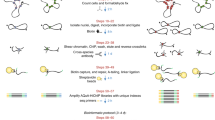
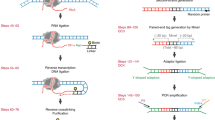
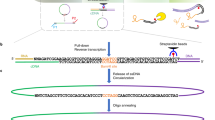
We present the experimental protocol and data analysis toolbox for multi-contact 4C (MC-4C), a new proximity ligation method tailored to study the higher-order chromatin contact patterns of selected genomic sites. Conventional chromatin conformation capture (3C) methods fragment proximity ligation products for efficient analysis of pairwise DNA contacts. By contrast, MC-4C is designed to preserve and collect large concatemers of proximity ligated fragments for long-molecule sequencing on an Oxford Nanopore or Pacific Biosciences platform. Each concatemer of proximity ligation products represents a snapshot topology of a different individual allele, revealing its multi-way chromatin interactions. By inverse PCR with primers specific for a fragment of interest (the viewpoint) and DNA size selection, sequencing is selectively targeted to thousands of different complex interactions containing this viewpoint. A tailored statistical analysis toolbox is able to generate background models and three-way interaction profiles from the same dataset. These profiles can be used to distinguish whether contacts between more than two regulatory sequences are mutually exclusive or, conversely, simultaneously occurring at chromatin hubs. The entire procedure can be completed in 2 w, and requires standard molecular biology and data analysis skills and equipment, plus access to a third-generation sequencing platform.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout














Similar content being viewed by others
Data availability
All raw sequencing data used in this study are available through the European Nucleotide Archive (https://www.ebi.ac.uk/ena/data/view/PRJEB23327). The processed data can be downloaded from the Mendeley Data repository (https://doi.org/10.17632/wbk8hk87r2.3). The test data used in the Walkthrough section of this protocol can be downloaded from github (https://github.com/deLaatLab/mc4c_test-data/archive/master.zip) and are included as Supplementary Data 2.
Code availability
The MC-4C pipeline (version 1.02, at the moment of protocol preparation), including all scripts used to produce figures in this protocol, is available under MIT License at github in mc4c_py repository (https://github.com/deLaatLab/mc4c_py) and is available directly as Supplementary Data 1. The data and results presented in this protocol were prepared using version 1.02 of the MC-4C pipeline.
References
Dekker, J., Rippe, K., Dekker, M. & Kleckner, N. Capturing chromosome conformation. Science 295, 1306–1311 (2002).
Denker, A. & de Laat, W. The second decade of 3C technologies: detailed insights into nuclear organization. Genes Dev. 30, 1357–1382 (2016).
Brant, L. et al. Exploiting native forces to capture chromosome conformation in mammalian cell nuclei. Mol. Syst. Biol. 12, 891 (2016).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Simonis, M. et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 (2006).
Splinter, E., de Wit, E., van de Werken, H. J., Klous, P. & de Laat, W. Determining long-range chromatin interactions for selected genomic sites using 4C-seq technology: from fixation to computation. Methods 58, 221–230 (2012).
Davies, J. O. et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat. Methods 13, 74–80 (2016).
Dostie, J. et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 (2006).
Li, G. et al. ChIA-PET tool for comprehensive chromatin interaction analysis with paired-end tag sequencing. Genome Biol. 11, R22 (2010).
Mumbach, M. R. et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016).
Fullwood, M. J. & Ruan, Y. ChIP-based methods for the identification of long-range chromatin interactions. J. Cell. Biochem. 107, 30–39 (2009).
Dixon, J. R. et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 (2012).
Nora, E. P. et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature 485, 381–385 (2012).
Sexton, T. et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell 148, 458–472 (2012).
Allahyar, A. et al. Enhancer hubs and loop collisions identified from single-allele topologies. Nat. Genet. 50, 1151–1160 (2018).
Bianco, S., Chiariello, A. M., Annunziatella, C., Esposito, A. & Nicodemi, M. Predicting chromatin architecture from models of polymer physics. Chromosome Res. 25, 25–34 (2017).
Serra, F. et al. Automatic analysis and 3D-modelling of Hi-C data using TADbit reveals structural features of the fly chromatin colors. PLoS Comput. Biol. 13, e1005665 (2017).
Olivares-Chauvet, P. et al. Capturing pairwise and multi-way chromosomal conformations using chromosomal walks. Nature 540, 296–300 (2016).
Darrow, E. M. et al. Deletion of DXZ4 on the human inactive X chromosome alters higher-order genome architecture. Proc. Natl Acad. Sci. USA 113, E4504–12 (2016).
Quinodoz, S. A. et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell 174, 744–757.e24 (2018).
Beagrie, R. A. et al. Complex multi-enhancer contacts captured by genome architecture mapping. Nature 543, 519–524 (2017).
Oudelaar, A. M. et al. Single-allele chromatin interactions identify regulatory hubs in dynamic compartmentalized domains. Nat. Genet. 50, 1744–1751 (2018).
van de Werken, H. J. et al. 4C technology: protocols and data analysis. Methods Enzymol. 513, 89–112 (2012).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 26, 589–595 (2010).
Koranne, S. Hierarchical data format 5: HDF5. in Handbook of Open Source Tools 191–200 (Springer, 2011).
Walt, S., van der, van der Walt, S., Chris Colbert, S. & Varoquaux, G. The NumPy array: a structure for efficient numerical computation. Comput. Sci. Eng. 13, 22–30 (2011).
McKinney, W. Python for Data Analysis: Data Wrangling with Pandas, NumPy, and IPython (‘O’Reilly Media, Inc.’, 2017).
Hunter, J. D. Matplotlib: a 2D graphics environment. Comput. Sci. Eng. 9, 90–95 (2007).
Nakayama, T. et al. Cas9-based genome editing in Xenopus tropicalis. Methods Enzymol. 546, 355–375 (2014).
Koh, C. M. Isolation of genomic DNA from mammalian cells. Methods Enzymol. 529, 161–169 (2013).
Acknowledgements
We thank E. Vos and E. deWit and other laboratory members for their input at various stages of the development of MC-4C. This work was supported by an NWO VIDI grant (639.072.715) to J.d.R and an NWO/CW TOP grant (714.012.002) and NWO VICI grant (724.012.003) to W.d.L., and by the NIH Common Fund Program, grant U01CA200147, as a Transformative Collaborative Project Award (TCPA; TCPA-2017-DE-LAAT).
Author information
Authors and Affiliations
Contributions
C.V. and B.A.M.B. designed and performed experiments. C.V. and A.A. wrote manuscript and designed the figures. A.A. designed and performed the computational analysis, designed the figures and wrote the manuscript. P.H.L.K., M.J.A.M.V. and C.V.-Q. performed ‘C’ methods experiments. R.S. and A.A. implemented the pipeline in Python. I.J.R. performed and W.P.K. designed and supervised MinION sequencing experiments. G.G. helped with computational analysis. J.d.R. designed and supervised the computational analyses and pipelines and co-wrote the manuscript. W.d.L. conceived and supervised the study and wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
C.V., B.A.M.B., P.H.L.K., M.J.A.M.V. and G.G. are shareholders of Cergentis. W.d.L. is founder and shareholder of Cergentis. W.P.K. and J.d.R. are co-founders and shareholders of Cyclomics.
Additional information
Peer review information Nature Protocols thanks Jian Ma and the other, anonymous, reviewer(s) for their contribution to the peer review of this work.
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key references using this protocol
Allahyar, A. et al. Nat. Genet. 50, 1151–1160 (2018): https://doi.org/10.1038/s41588-018-0161-5
Integrated supplementary information
Supplementary Fig. 1 gRNA template.
5 μl of gRNA template PCR was loaded on a 2% agarose gel stained with ethidium bromide.
Supplementary information
Supplementary Information
Supplementary Fig. 1 and Supplementary Manual
Supplementary Data 1
MC-4C pipeline
Supplementary Data 2
MC-4C test dataset
Rights and permissions
About this article
Cite this article
Vermeulen, C., Allahyar, A., Bouwman, B.A.M. et al. Multi-contact 4C: long-molecule sequencing of complex proximity ligation products to uncover local cooperative and competitive chromatin topologies. Nat Protoc 15, 364–397 (2020). https://doi.org/10.1038/s41596-019-0242-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0242-7
This article is cited by
-
Beyond assembly: the increasing flexibility of single-molecule sequencing technology
Nature Reviews Genetics (2023)
-
Capture-C: a modular and flexible approach for high-resolution chromosome conformation capture
Nature Protocols (2022)
-
SPRITE: a genome-wide method for mapping higher-order 3D interactions in the nucleus using combinatorial split-and-pool barcoding
Nature Protocols (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.