Abstract
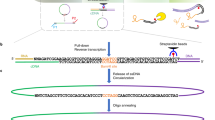
Chromatin in higher eukaryotic nuclei is extensively bound by various RNA species. We recently developed a method for in situ capture of global RNA interactions with DNA by deep sequencing (GRID-seq) of fixed permeabilized nuclei that allows identification of the entire repertoire of chromatin-associated RNAs in an unbiased manner. The experimental design of GRID-seq is related to those of two recently published strategies (MARGI (mapping RNA–genome interactions) and ChAR-seq (chromatin-associated RNA sequencing)), which also use a bivalent linker to ligate RNA and DNA in proximity. Importantly, however, GRID-seq also implements a combined experimental and computational approach to control nonspecific RNA–DNA interactions that are likely to occur during library construction, which is critical for accurate interpretation of detected RNA–DNA interactions. GRID-seq typically finds both coding and non-coding RNAs (ncRNAs) that interact with tissue-specific promoters and enhancers, especially super-enhancers, from which a global promoter–enhancer connectivity map can be deduced. Here, we provide a detailed protocol for GRID-seq that includes nuclei preparation, chromatin fragmentation, RNA and DNA in situ ligation with a bivalent linker, PCR amplification and high-throughput sequencing. To further enhance the utility of GRID-seq, we include a pipeline for data analysis, called GridTools, into which key steps such as background correction and inference of genomic element proximity are integrated. For researchers experienced in molecular biology with minimal bioinformatics skills, the protocol typically takes 4–5 d from cell fixation to library construction and 2–3 d for data processing.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Data availability
Data obtained with B6 mESCs have been deposited in the Gene Expression Omnibus under accession no. GSE82312, and a test dataset on the F123 mESCs is accessible at http://fugenome.ucsd.edu/gridseq/datasets/gridseq.test10M.raw.fq.gz.
Code availability
The in-house software, GridTools, is publicly available in GitHub at https://github.com/biz007/gridtools under an MIT license. It also houses a demo code for the F123 mESC test dataset. All analyses of the results and the performance evaluation of the pipeline in the current study were based on GridTools v.1.1 and are also available as Supplementary Software.
Change history
14 February 2020
An amendment to this paper has been published and can be accessed via a link at the top of the paper.
References
Djebali, S. et al. Landscape of transcription in human cells. Nature 489, 101–108 (2012).
Fu, X. D. Non-coding RNA: a new frontier in regulatory biology. Natl. Sci. Rev. 1, 190–204 (2014).
Rinn, J. L. & Chang, H. Y. Genome regulation by long noncoding RNAs. Annu. Rev. Biochem. 81, 145–166 (2012).
Nguyen, T. C., Zaleta-Rivera, K., Huang, X., Dai, X. & Zhong, S. RNA, action through interactions. Trends Genet. 34, 867–882 (2018).
Chu, C., Qu, K., Zhong, F. L., Artandi, S. E. & Chang, H. Y. Genomic maps of long noncoding RNA occupancy reveal principles of RNA–chromatin interactions. Mol. Cell 44, 667–678 (2011).
Simon, M. D. et al. The genomic binding sites of a noncoding RNA. Proc. Natl. Acad. Sci. USA 108, 20497–20502 (2011).
Engreitz, J. M. et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 341, 1237973 (2013).
Li, X. et al. GRID-seq reveals the global RNA–chromatin interactome. Nat. Biotechnol. 35, 940–950 (2017).
Sridhar, B. et al. Systematic mapping of RNA–chromatin interactions in vivo. Curr. Biol. 27, 602–609 (2017).
Bell, J.C. et al. Chromatin-associated RNA sequencing (ChAR-seq) maps genome-wide RNA-to-DNA contacts. Elife 7, e27024 (2018).
Mumbach, M. R. et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture. Nat. Methods 13, 919–922 (2016).
Fang, R. et al. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 26, 1345–1348 (2016).
Whyte, W. A. et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell 153, 307–319 (2013).
Lieberman-Aiden, E. et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 (2009).
Rao, S. S. et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell 159, 1665–1680 (2014).
Saha, S. et al. Using the transcriptome to annotate the genome. Nat. Biotechnol. 20, 508–512 (2002).
Fullwood, M. J. et al. An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462, 58–64 (2009).
Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 3–12 (2011).
Li, H. & Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 26, 589–595 (2010).
Li, H. et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Acknowledgements
We thank B. Ren (University of California, San Diego) for the generous gifts of C57BL/6 mouse ES cells and F1 Mus musculus castaneus × S129/SvJae mouse ES cells. This work was supported by NIH grants (HG004659, HG007005, GM049369 and DK098808) to X.-D.F.; a grant from the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010113) to B.Z.; grants from the National Science Foundation of China (31872191, 31472263) and the China Scholarship Council (201606275037) to D.L.; and a grant from the National Natural Science Foundation of China (91640115) to Y.Z.
Author information
Authors and Affiliations
Contributions
X.L. and X.-D.F. designed and implemented the original GRID-seq experimental protocol; D.L. and D.-H.L. optimized the protocol. B.Z., X.L. and X.-D.F. designed the data-processing pipeline and B.Z. designed the GridTools software with assistance from Y.Z.; X.L., B.Z., D.L., D.-H.L. and X.-D.F. prepared the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Journal peer review information: Nature Protocols thanks Xiaohua Shen and other anonymous reviewer(s) for their contribution to the peer review of this work.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related link
Key reference using this protocol
Li, X. et al. Nat. Biotech. 35, 940–950 (2017): https://doi.org/10.1038/nbt.3968
Integrated supplementary information
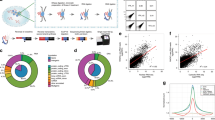
Supplementary Fig. 1 Identification of chromatin-associated RNAs.
(a) All mouse genes were ranked in a descending fashion by their normalized RNA read densities captured by GRID-seq in B6 mESC datasets. Each dot represents one gene and red ones highlight genes with the RNA read density of >100 RPK. RPK: reads per kb. (b) Similar plot based on F123 mESC datasets as in a. (c) All genes were ranked in a descending fashion by the maximal linked DNA read density on all genomic bins in B6 mESC datasets. Each dot represents one gene and blue ones emphasize genes with DNA read density of >10 RPK. (d) Similar plot based on F123 mESC datasets as in c. (e) Scatter plot showing the distribution of all genes based on their RNA read densities (x-axis) and maximal DNA read densities over genomic bins (y-axis) in B6 mESC datasets. Red dots correspond to genes marked with red dots in a; blue dots correspond to genes marked with blue dots in c; purple dots represent chromatin-associated RNA that survived both cutoffs. (f) Similar plot based on F123 mESC datasets as in e. (g, h) Saturation analyses performed by random subsampling 5 to 55 million uniquely mapped read mates in B6 and 5 to 35 million uniquely mapped read mates in F123 mESC datasets. Each level of random subsampling was independently repeated 5 times. Numbers of chromatin-associated RNAs identified were summarized and plotted in g. The green line represents the connected mean values in B6, and the orange dotted line represents those in the F123 mESC dataset. Numbers of 1 kb-binned genomic regions that have at least 10 linked DNA reads were summarized and plotted in h. The blue line represents the connected mean values in B6, and the red dotted line represents those in the F123 mESC dataset.
Supplementary Fig. 2 Reproducibility of GRID-seq libraries.
(a) Smoothed scatter plots of uniquely mapped RNA read levels for all genes between replicated libraries of B6 (left) and F123 mESCs (right). (b) Smoothed scatter plots of uniquely mapped DNA read levels for all 10-kb genomic bins between replicated libraries of B6 (left) and F123 mESCs (right). (c) Smoothed scatter plots of background signals over all 10-kb genomic bins (B values) between replicated libraries of B6 (left) and F123 mESC (right). (d) Smoothed scatter plots of foreground signals over all 10-kb genomic bins (V values) between replicated libraries in B6 (left) and F123 mESC (right). (e) Overlapping of chromatin-associated RNAs identified in replicated and combined B6 mESC datasets. The numbers on the plot indicate the chromatin-associated RNA species in respectively coloured areas in the Venn diagram (for example: 505 common chromatin-associated RNAs are identified in the combined, replicate 1 and replicate 2 datasets; 13 chromatin-associated RNAs are identified in the combined and the replicate 2 data but not in the replicate 1 data). (f) Overlapping of chromatin-associated RNAs identified in replicated and combined F123 mESC datasets, displayed in a similar manner as in e. (g) RNA–chromatin interactome on chromosome 11 independently constructed from B6 mESC replicates. Boxed regions in each panel were enlarged with increasing resolution in the next panel on the right. A set of representative chromatin-associated RNAs are labeled on the right. (h) Interactome on chromosome 2 independently constructed from F123 mESC replicates similar to g.
Supplementary information
Rights and permissions
About this article
Cite this article
Zhou, B., Li, X., Luo, D. et al. GRID-seq for comprehensive analysis of global RNA–chromatin interactions. Nat Protoc 14, 2036–2068 (2019). https://doi.org/10.1038/s41596-019-0172-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41596-019-0172-4
This article is cited by
-
From ‘Omics to Multi-omics Technologies: the Discovery of Novel Causal Mediators
Current Atherosclerosis Reports (2023)
-
Long non-coding RNAs: definitions, functions, challenges and recommendations
Nature Reviews Molecular Cell Biology (2023)
-
Global profiling of RNA–chromatin interactions reveals co-regulatory gene expression networks in Arabidopsis
Nature Plants (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.