Abstract
The dynamic and polymicrobial oral microbiome is a direct precursor of diseases such as dental caries and periodontitis, two of the most prevalent microbially induced disorders worldwide. Distinct microenvironments at oral barriers harbour unique microbial communities, which are regulated through sophisticated signalling systems and by host and environmental factors. The collective function of microbial communities is a major driver of homeostasis or dysbiosis and ultimately health or disease. Despite different aetiologies, periodontitis and caries are each driven by a feedforward loop between the microbiota and host factors (inflammation and dietary sugars, respectively) that favours the emergence and persistence of dysbiosis. In this Review, we discuss current knowledge and emerging mechanisms governing oral polymicrobial synergy and dysbiosis that have both enhanced our understanding of pathogenic mechanisms and aided the design of innovative therapeutic approaches for oral diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$29.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


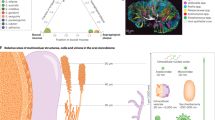
Part a adapted with permission from ref.56, Wiley-VCH.

Adapted with permission from ref.7, Cell Press.



Adapted with permission from ref.10, Cell Press.
Similar content being viewed by others
References
Abusleme, L. et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. ISME J. 7, 1016–1025 (2013). This study documents alterations in subgingival microbial communities that underpin the development of periodontitis and describes the relationship between clinical inflammation and the disease-associated microbiome.
Griffen, A. L. et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. ISME J. 6, 1176–1185 (2012). This landmark study establishes the complexity of the periodontal microbial community and the demarcation between health and disease.
Rosan, B. & Lamont, R. J. Dental plaque formation. Microbes Infect. 2, 1599–1607 (2000).
Aas, J. A., Paster, B. J., Stokes, L. N., Olsen, I. & Dewhirst, F. E. Defining the normal bacterial flora of the oral cavity. J. Clin. Microbiol. 43, 5721–5732 (2005).
Dewhirst, F. E. et al. The human oral microbiome. J. Bacteriol. 192, 5002–5017 (2010). References 4 and 5 are the basis of our current understanding of the diversity of the oral microbiome.
Mark Welch, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E. & Borisy, G. G. Biogeography of a human oral microbiome at the micron scale. Proc. Natl Acad. Sci. USA 113, E791–E800 (2016). This imaging study provides the foundation for concepts of oral microbial biogeography.
Bowen, W. H., Burne, R. A., Wu, H. & Koo, H. Oral biofilms: pathogens, matrix, and polymicrobial interactions in microenvironments. Trends Microbiol. 26, 229–242 (2018).
Kamada, N., Chen, G. Y., Inohara, N. & Nunez, G. Control of pathogens and pathobionts by the gut microbiota. Nat. Immunol. 14, 685–690 (2013).
Kamada, N., Seo, S. U., Chen, G. Y. & Nunez, G. Role of the gut microbiota in immunity and inflammatory disease. Nat. Rev. Immunol. 13, 321–335 (2013).
Lamont, R. J. & Hajishengallis, G. Polymicrobial synergy and dysbiosis in inflammatory disease. Trends Mol. Med. 21, 172–183 (2015).
Hajishengallis, G. & Lamont, R. J. Breaking bad: manipulation of the host response by Porphyromonas gingivalis. Eur. J. Immunol. 44, 328–338 (2014).
Baker, J. L., Bor, B., Agnello, M., Shi, W. & He, X. Ecology of the oral microbiome: beyond bacteria. Trends Microbiol. 25, 362–374 (2017).
Dabdoub, S. M., Ganesan, S. M. & Kumar, P. S. Comparative metagenomics reveals taxonomically idiosyncratic yet functionally congruent communities in periodontitis. Sci. Rep. 6, 38993 (2016).
Suwannakul, S., Stafford, G. P., Whawell, S. A. & Douglas, C. W. Identification of bistable populations of Porphyromonas gingivalis that differ in epithelial cell invasion. Microbiology 156, 3052–3064 (2010).
Valm, A. M. et al. Systems-level analysis of microbial community organization through combinatorial labeling and spectral imaging. Proc. Natl Acad. Sci. USA 108, 4152–4157 (2011).
Marsh, P. D. & Zaura, E. Dental biofilm: ecological interactions in health and disease. J. Clin. Periodontol. 44 (Suppl. 18), 12–22 (2017).
Takahashi, N. & Nyvad, B. The role of bacteria in the caries process: ecological perspectives. J. Dent. Res. 90, 294–303 (2011).
Xiao, J. et al. The exopolysaccharide matrix modulates the interaction between 3D architecture and virulence of a mixed-species oral biofilm. PLOS Pathog. 8, e1002623 (2012).
Guo, L., McLean, J. S., Lux, R., He, X. & Shi, W. The well-coordinated linkage between acidogenicity and aciduricity via insoluble glucans on the surface of Streptococcus mutans. Sci. Rep. 5, 18015 (2015).
Hajishengallis, E., Parsaei, Y., Klein, M. I. & Koo, H. Advances in the microbial etiology and pathogenesis of early childhood caries. Mol. Oral Microbiol. 32, 24–34 (2017).
Mira, A., Simon-Soro, A. & Curtis, M. A. Role of microbial communities in the pathogenesis of periodontal diseases and caries. J. Clin. Periodontol. 44 (Suppl. 18), S23–S38 (2017).
Tanner, A. C. R., Kressirer, C. A., Rothmiller, S., Johansson, I. & Chalmers, N. I. The caries microbiome: implications for reversing dysbiosis. Adv. Dent. Res. 29, 78–85 (2018).
Eriksson, L., Lif Holgerson, P., Esberg, A. & Johansson, I. Microbial complexes and caries in 17-year-olds with and without Streptococcus mutans. J. Dent. Res. 97, 275–282 (2018).
Hajishengallis, G. & Lamont, R. J. Beyond the red complex and into more complexity: the polymicrobial synergy and dysbiosis (PSD) model of periodontal disease etiology. Mol. Oral Microbiol. 27, 409–419 (2012). This paper questions the primary importance of individual pathogens, such as the red complex bacteria, and proposes that periodontitis is initiated by a synergistic polymicrobial community within which different species, or specific gene combinations thereof, mediate distinct roles that converge to shape and stabilize a dysbiotic and disease-provoking microbiota.
Holt, S. C. & Ebersole, J. L. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol. 2000 38, 72–122 (2005).
Socransky, S. S. & Haffajee, A. D. Periodontal microbial ecology. Periodontol. 2000 38, 135–187 (2005).
Dewhirst, F. E. The oral microbiome: critical for understanding oral health and disease. J. Calif. Dent. Assoc. 44, 409–410 (2016).
Diaz, P. I., Hoare, A. & Hong, B. Y. Subgingival microbiome shifts and community dynamics in periodontal diseases. J. Calif. Dent. Assoc. 44, 421–435 (2016).
Moore, W. E. et al. The microflora of periodontal sites showing active destructive progression. J. Clin. Periodontol. 18, 729–739 (1991).
Diaz, P. I. Microbial diversity and interactions in subgingival biofilm communities. Front. Oral Biol. 15, 17–40 (2012).
Simon-Soro, A. et al. Microbial geography of the oral cavity. J. Dent. Res. 92, 616–621 (2013).
Nowicki, E. M. et al. Microbiota and metatranscriptome changes accompanying the onset of gingivitis. mBio 9, e00575–18 (2018).
Whitmore, S. E. & Lamont, R. J. Oral bacteria and cancer. PLOS Pathog. 10, e1003933 (2014).
Atanasova, K. R. & Yilmaz, O. Looking in the Porphyromonas gingivalis cabinet of curiosities: the microbium, the host and cancer association. Mol. Oral Microbiol. 29, 55–66 (2014).
Sahingur, S. E. & Yeudall, W. A. Chemokine function in periodontal disease and oral cavity cancer. Front. Immunol. 6, 214 (2015).
Cugini, C., Klepac-Ceraj, V., Rackaityte, E., Riggs, J. E. & Davey, M. E. Porphyromonas gingivalis: keeping the pathos out of the biont. J. Oral Microbiol. 5, 19804 (2013).
Takahashi, N. Oral microbiome metabolism: from “who are they?” to “what are they doing?”. J. Dent. Res. 94, 1628–1637 (2015).
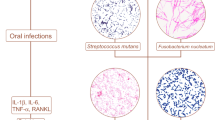
Han, Y. W. & Wang, X. Mobile microbiome: oral bacteria in extra-oral infections and inflammation. J. Dent. Res. 92, 485–491 (2013).
Hajishengallis, G. Periodontitis: from microbial immune subversion to systemic inflammation. Nat. Rev. Immunol. 15, 30–44 (2015).
Kumar, P. S. Oral microbiota and systemic disease. Anaerobe 24, 90–93 (2013).
Maddi, A. & Scannapieco, F. A. Oral biofilms, oral and periodontal infections, and systemic disease. Am. J. Dent. 26, 249–254 (2013).
Potempa, J., Mydel, P. & Koziel, J. The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat. Rev. Rheumatol. 13, 606–620 (2017).
Ebersole, J. L. et al. The periodontal war: microbes and immunity. Periodontol. 2000 75, 52–115 (2017).
Chukkapalli, S. S. et al. Global TLR2 and 4 deficiency in mice impacts bone resorption, inflammatory markers and atherosclerosis to polymicrobial infection. Mol. Oral Microbiol. 32, 211–225 (2017).
Hajishengallis, G. & Lamont, R. J. Dancing with the stars: how choreographed bacterial interactions dictate nososymbiocity and give rise to keystone pathogens, accessory pathogens, and pathobionts. Trends Microbiol. 24, 477–489 (2016).
Murray, J. L., Connell, J. L., Stacy, A., Turner, K. H. & Whiteley, M. Mechanisms of synergy in polymicrobial infections. J. Microbiol. 52, 188–199 (2014).
Michie, K. L., Cornforth, D. M. & Whiteley, M. Bacterial tweets and podcasts #signaling#eavesdropping#microbialfightclub. Mol. Biochem. Parasitol. 208, 41–48 (2016).
Short, F. L., Murdoch, S. L. & Ryan, R. P. Polybacterial human disease: the ills of social networking. Trends Microbiol. 22, 508–516 (2014).
Stacy, A., McNally, L., Darch, S. E., Brown, S. P. & Whiteley, M. The biogeography of polymicrobial infection. Nat. Rev. Microbiol. 14, 93–105 (2016).
Hwang, G. et al. Candida albicans mannans mediate Streptococcus mutans exoenzyme GtfB binding to modulate cross-kingdom biofilm development in vivo. PLOS Pathog. 13, e1006407 (2017).
Kim, D. et al. Candida albicans stimulates Streptococcus mutans microcolony development via cross-kingdom biofilm-derived metabolites. Sci. Rep. 7, 41332 (2017).
Chen, Y. E., Fischbach, M. A. & Belkaid, Y. Skin microbiota-host interactions. Nature 553, 427–436 (2018).
Casadevall, A. The pathogenic potential of a microbe. mSphere 2, e00015–17 (2017).
Lin, D. & Koskella, B. Friend and foe: factors influencing the movement of the bacterium Helicobacter pylori along the parasitism-mutualism continuum. Evol. Appl. 8, 9–22 (2015).
Nelson, P. G. & May, G. Coevolution between mutualists and parasites in symbiotic communities may lead to the evolution of lower virulence. Am. Nat. 190, 803–817 (2017).
Whitmore, S. E. & Lamont, R. J. The pathogenic persona of community-associated oral streptococci. Mol. Microbiol. 81, 305–314 (2011).
Nobbs, A. H., Lamont, R. J. & Jenkinson, H. F. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73, 407–450 (2009).
Lee, S. F. Oral colonization and immune responses to Streptococcus gordonii: potential use as a vector to induce antibodies against respiratory pathogens. Curr. Opin. Infect. Dis. 16, 231–235 (2003).
Xie, E. et al. Oral delivery of a novel recombinant Streptococcus mitis vector elicits robust vaccine antigen-specific oral mucosal and systemic antibody responses and T cell tolerance. PLOS ONE 10, e0143422 (2015).
Daep, C. A., Novak, E. A., Lamont, R. J. & Demuth, D. R. Structural dissection and in vivo effectiveness of a peptide inhibitor of Porphyromonas gingivalis adherence to Streptococcus gordonii. Infect. Immun. 79, 67–74 (2011).
Kuboniwa, M. et al. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. Nat. Microbiol. 2, 1493–1499 (2017). This study establishes multidimensional communication between two organisms in the oral community that separately either enhance or suppress nososymbiocity.
Wright, C. J. et al. Characterization of a bacterial tyrosine kinase in Porphyromonas gingivalis involved in polymicrobial synergy. Microbiologyopen 3, 383–394 (2014).
Liu, C., Miller, D. P., Wang, Y., Merchant, M. & Lamont, R. J. Structure-function aspects of the Porphyromonas gingivalis tyrosine kinase Ptk1. Mol. Oral Microbiol. 32, 314–323 (2017).
Ramsey, M. M. & Whiteley, M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proc. Natl Acad. Sci. USA 106, 1578–1583 (2009).
Stacy, A. et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proc. Natl Acad. Sci. USA 111, 7819–7824 (2014). This study reveals that the spatial proximity of organisms can depend on the collective outcome of synergistic and antagonistic interactions.
Duan, D., Scoffield, J. A., Zhou, X. & Wu, H. Fine-tuned production of hydrogen peroxide promotes biofilm formation of Streptococcus parasanguinis by a pathogenic cohabitant Aggregatibacter actinomycetemcomitans. Environ. Microbiol. 18, 4023–4036 (2016).
Bertolini, M. M. et al. Candida-streptococcal mucosal biofilms display distinct structural and virulence characteristics depending on growth conditions and hyphal morphotypes. Mol. Oral Microbiol. 30, 307–322 (2015).
Xu, H., Jenkinson, H. F. & Dongari-Bagtzoglou, A. Innocent until proven guilty: mechanisms and roles of Streptococcus-Candida interactions in oral health and disease. Mol. Oral Microbiol. 29, 99–116 (2014).
Cheng, X. et al. Plasticity of the pyruvate node modulates hydrogen peroxide production and acid tolerance in multiple oral Streptococci. Appl. Environ. Microbiol. 84, e01697–17 (2018).
Thurnheer, T. & Belibasakis, G. N. Streptococcus oralis maintains homeostasis in oral biofilms by antagonizing the cariogenic pathogen Streptococcus mutans. Mol. Oral Microbiol. 33, 234–239 (2018).
Redanz, S. et al. Live and let die: hydrogen peroxide production by the commensal flora and its role in maintaining a symbiotic microbiome. Mol. Oral Microbiol. https://doi.org/10.1111/omi.12231 (2018).
Ho, M. H., Lamont, R. J. & Xie, H. Identification of Streptococcus cristatus peptides that repress expression of virulence genes in Porphyromonas gingivalis. Sci. Rep. 7, 1413 (2017).
Ho, M. H., Lamont, R. J. & Xie, H. A novel peptidic inhibitor derived from Streptococcus cristatus ArcA attenuates virulence potential of Porphyromonas gingivalis. Sci. Rep. 7, 16217 (2017).
Xie, H., Hong, J., Sharma, A. & Wang, B. Y. Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. J. Periodontal. Res. 47, 578–583 (2012).
Wang, B. Y., Wu, J., Lamont, R. J., Lin, X. & Xie, H. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J. Clin. Microbiol. 47, 3902–3906 (2009).
Mans, J. J., Hendrickson, E. L., Hackett, M. & Lamont, R. J. Cellular and bacterial profiles associated with oral epithelium-microbiota interactions. Periodontol. 2000 52, 207–217 (2010).
Colombo, A. V., Silva, C. M., Haffajee, A. & Colombo, A. P. Identification of oral bacteria associated with crevicular epithelial cells from chronic periodontitis lesions. J. Med. Microbiol. 55, 609–615 (2006).
Kreth, J., Giacaman, R. A., Raghavan, R. & Merritt, J. The road less traveled - defining molecular commensalism with Streptococcus sanguinis. Mol. Oral Microbiol. 32, 181–196 (2017).
Shah, S. A. et al. The making of a miscreant: tobacco smoke and the creation of pathogen-rich biofilms. NPJ Biofilms Microbiomes 3, 26 (2017).
Ghosh, S. K. et al. Conceptual perspectives: bacterial antimicrobial peptide induction as a novel strategy for symbiosis with the human host. Front. Microbiol. 9, 302 (2018).
Al-Attar, A. et al. Activation of Notch-1 in oral epithelial cells by P. gingivalis triggers the expression of the antimicrobial protein PLA2-IIA. Mucosal Immunol. 11, 1047–1059 (2018).
Pitts, N. B. et al. Dental caries. Nat. Rev. Dis. Primers 3, 17030 (2017).
Liu, Y. L., Nascimento, M. & Burne, R. A. Progress toward understanding the contribution of alkali generation in dental biofilms to inhibition of dental caries. Int. J. Oral Sci. 4, 135–140 (2012).
Merritt, J. & Qi, F. The mutacins of Streptococcus mutans: regulation and ecology. Mol. Oral Microbiol. 27, 57–69 (2012).
Qi, F. & Kreth, J. Methods to study antagonistic activities among oral bacteria. Methods Mol. Biol. 1537, 203–218 (2017).
Gross, E. L. et al. Beyond Streptococcus mutans: dental caries onset linked to multiple species by 16S rRNA community analysis. PLOS ONE 7, e47722 (2012).
Johansson, I., Witkowska, E., Kaveh, B., Lif Holgerson, P. & Tanner, A. C. The microbiome in populations with a low and high prevalence of caries. J. Dent. Res. 95, 80–86 (2016).
Teng, F. et al. Prediction of early childhood caries via spatial-temporal variations of oral microbiota. Cell Host Microbe 18, 296–306 (2015).
Takahashi, N. & Nyvad, B. Ecological hypothesis of dentin and root caries. Caries Res. 50, 422–431 (2016).
Richards, V. P. et al. Microbiomes of site-specific dental plaques from children with different caries status. Infect. Immun. 85, e00106–17 (2017).
Knapp, S. et al. Natural competence is common among clinical isolates of Veillonella parvula and is useful for genetic manipulation of this key member of the oral microbiome. Front. Cell. Infect. Microbiol. 7, 139 (2017).
Mashima, I. & Nakazawa, F. The influence of oral Veillonella species on biofilms formed by Streptococcus species. Anaerobe 28, 54–61 (2014).
Xiao, J. et al. Candida albicans and early childhood caries: a systematic review and meta-analysis. Caries Res. 52, 102–112 (2018).
Falsetta, M. L. et al. Symbiotic relationship between Streptococcus mutans and Candida albicans synergizes virulence of plaque biofilms in vivo. Infect. Immun. 82, 1968–1981 (2014).
Sztajer, H. et al. Cross-feeding and interkingdom communication in dual-species biofilms of Streptococcus mutans and Candida albicans. ISME J. 8, 2256–2271 (2014).
Flemming, H. C. et al. Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575 (2016).
Hwang, G. et al. Simultaneous spatiotemporal mapping of in situ pH and bacterial activity within an intact 3D microcolony structure. Sci. Rep. 6, 32841 (2016).
Jiang, W. et al. Pyrosequencing analysis of oral microbiota shifting in various caries states in childhood. Microb. Ecol. 67, 962–969 (2014).
Marsh, P. D. Are dental diseases examples of ecological catastrophes? Microbiology 149, 279–294 (2003). This paper proposes that environmental factors drive the selection and enrichment of specific oral pathogenic bacteria, with implications for both dental caries and periodontitis.
Hasturk, H. et al. Resolvin E1 regulates inflammation at the cellular and tissue level and restores tissue homeostasis in vivo. J. Immunol. 179, 7021–7029 (2007).
Abe, T. et al. Local complement-targeted intervention in periodontitis: proof-of-concept using a C5a receptor (CD88) antagonist. J. Immunol. 189, 5442–5448 (2012).
Hajishengallis, G. et al. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe 10, 497–506 (2011). This study substantiates the concept of a keystone pathogen by providing in vivo evidence that a specific microorganism instigates quantitative and qualitative alterations to the commensal microbiota, which is thereby remodelled into a dysbiotic community driving periodontitis.
Eskan, M. A. et al. The leukocyte integrin antagonist Del-1 inhibits IL-17-mediated inflammatory bone loss. Nat. Immunol. 13, 465–473 (2012).
Moutsopoulos, N. M. et al. Defective neutrophil recruitment in leukocyte adhesion deficiency type I disease causes local IL-17–driven inflammatory bone loss. Sci. Transl Med. 6, 229ra240 (2014).
Lee, C.-T. et al. Resolvin E1 reverses experimental periodontitis and dysbiosis. J. Immunol. 197, 2796–2806 (2016).
Hajishengallis, G. The inflammophilic character of the periodontitis-associated microbiota. Mol. Oral Microbiol. 29, 248–257 (2014).
Duran-Pinedo, A. E. et al. Community-wide transcriptome of the oral microbiome in subjects with and without periodontitis. ISME J. 8, 1659–1672 (2014).
Herrero, E. R. et al. Dysbiotic biofilms deregulate the periodontal inflammatory response. J. Dent. Res. 97, 547–555 (2018).
Yost, S., Duran-Pinedo, A. E., Teles, R., Krishnan, K. & Frias-Lopez, J. Functional signatures of oral dysbiosis during periodontitis progression revealed by microbial metatranscriptome analysis. Genome Med. 7, 27 (2015).
Bang, J., Cimasoni, G., Rosenbusch, C. & Duckert, A. Sodium, potassium and calcium contents of crevicular exudate: their relations to gingivitis and periodontitis. J. Periodontol. 44, 770–774 (1973).
Yost, S., Duran-Pinedo, A. E., Krishnan, K. & Frias-Lopez, J. Potassium is a key signal in host-microbiome dysbiosis in periodontitis. PLOS Pathog. 13, e1006457 (2017). This investigation identifies key metabolic changes in the periodontal microbial community associated with dysbiosis initiation and concludes that disease progression is mediated by the collective virulence of the entire community rather than by the action of a select few pathogens.
Winter, S. E. & Baumler, A. J. Dysbiosis in the inflamed intestine: chance favors the prepared microbe. Gut Microbes 5, 71–73 (2014).
Nassar, M. et al. GAS6 is a key homeostatic immunological regulator of host-commensal interactions in the oral mucosa. Proc. Natl Acad. Sci. USA 114, E337–E346 (2017).
Dalal, S. R. & Chang, E. B. The microbial basis of inflammatory bowel diseases. J. Clin. Invest. 124, 4190–4196 (2014).
Hajishengallis, G., Darveau, R. P. & Curtis, M. A. The keystone-pathogen hypothesis. Nat. Rev. Microbiol. 10, 717–725 (2012).
Finlay, B. B. & McFadden, G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell 124, 767–782 (2006).
Maekawa, T. et al. Porphyromonas gingivalis manipulates complement and TLR signaling to uncouple bacterial clearance from inflammation and promote dysbiosis. Cell Host Microbe 15, 768–778 (2014). This study shows that a keystone periodontal pathogen manipulates complement–TLR crosstalk to block bactericidal mechanisms while fostering a nutritionally favourable inflammatory response; this uncoupling of immune bacterial clearance from inflammation promotes dysbiosis and periodontitis.
Makkawi, H. et al. Porphyromonas gingivalis stimulates TLR2-PI3K signaling to escape immune clearance and induce bone resorption independently of MyD88. Front. Cell. Infect. Microbiol. 7, 359 (2017).
Wang, M. et al. Microbial hijacking of complement-toll-like receptor crosstalk. Sci. Signal. 3, ra11 (2010).
Liang, S. et al. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J. Immunol. 186, 869–877 (2011).
Tonetti, M. S., Cortellini, D. & Lang, N. P. In situ detection of apoptosis at sites of chronic bacterially induced inflammation in human gingiva. Infect. Immun. 66, 5190–5195 (1998).
Zenobia, C. et al. Commensal bacteria-dependent select expression of CXCL2 contributes to periodontal tissue homeostasis. Cell. Microbiol. 15, 1419–1426 (2013).
Darveau, R. P., Belton, C. M., Reife, R. A. & Lamont, R. J. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 66, 1660–1665 (1998).
Jauregui, C. E. et al. Suppression of T cell chemokines by Porphyromonas gingivalis. Infect. Immun. 81, 2288–2295 (2013).
Takeuchi, H. et al. The serine phosphatase SerB of Porphyromonas gingivalis suppresses IL-8 production by dephosphorylation of NF-kappaB RelA/p65. PLOS Pathog. 9, e1003326 (2013). References 123–125 establish the concept of P. gingivalis-induced local ‘chemokine paralysis’, originally shown to affect innate immunity and later expanded to include suppression of T cell-specific chemokines.
Hajishengallis, G. Immunomicrobial pathogenesis of periodontitis: keystones, pathobionts, and host response. Trends Immunol. 35, 3–11 (2014).
Moutsopoulos, N. M. et al. Subgingival microbial communities in leukocyte adhesion deficiency and their relationship with local immunopathology. PLOS Pathog. 11, e1004698 (2015).
Darveau, R. P., Hajishengallis, G. & Curtis, M. A. Porphyromonas gingivalis as a potential community activist for disease. J. Dent. Res. 91, 816–820 (2012).
Koo, H., Allan, R. N., Howlin, R. P., Stoodley, P. & Hall-Stoodley, L. Targeting microbial biofilms: current and prospective therapeutic strategies. Nat. Rev. Microbiol. 15, 740–755 (2017).
Liu, Y., Ren, Z., Hwang, G. & Koo, H. Therapeutic strategies targeting cariogenic biofilm microenvironment. Adv. Dent. Res. 29, 86–92 (2018).
Marquis, R. E., Clock, S. A. & Mota-Meira, M. Fluoride and organic weak acids as modulators of microbial physiology. FEMS Microbiol. Rev. 26, 493–510 (2003).
Kolderman, E. et al. L-Arginine destabilizes oral multi-species biofilm communities developed in human saliva. PLOS ONE 10, e0121835 (2015).
Nascimento, M. M. et al. The effect of arginine on oral biofilm communities. Mol. Oral Microbiol. 29, 45–54 (2014).
Pleszczynska, M., Wiater, A., Janczarek, M. & Szczodrak, J. (1-→3)-alpha-D-Glucan hydrolases in dental biofilm prevention and control: a review. Int. J. Biol. Macromol. 79, 761–778 (2015).
Fabbri, S. et al. High-velocity microsprays enhance antimicrobial activity in Streptococcus mutans biofilms. J. Dent. Res. 95, 1494–1500 (2016).
Liu, Y. et al. Topical delivery of low-cost protein drug candidates made in chloroplasts for biofilm disruption and uptake by oral epithelial cells. Biomaterials 105, 156–166 (2016).
Gao, L. et al. Nanocatalysts promote Streptococcus mutans biofilm matrix degradation and enhance bacterial killing to suppress dental caries in vivo. Biomaterials 101, 272–284 (2016).
Horev, B. et al. pH-activated nanoparticles for controlled topical delivery of farnesol to disrupt oral biofilm virulence. ACS Nano 9, 2390–2404 (2015).
Liu, Y. et al. Surface-adaptive, antimicrobially loaded, micellar nanocarriers with enhanced penetration and killing efficiency in Staphylococcal biofilms. ACS Nano 10, 4779–4789 (2016). References 137–139 demonstrate the feasibility of using pH-activated nanotechnologies for enhanced drug delivery, release or activation within the biofilm microenvironment to amplify the precision and efficacy of antibiofilm effects.
Paula, A. J. & Koo, H. Nanosized building blocks for customizing novel antibiofilm approaches. J. Dent. Res. 96, 128–136 (2017).
Maekawa, T. et al. Inhibition of pre-existing natural periodontitis in non-human primates by a locally administered peptide inhibitor of complement C3. J. Clin. Periodontol. 43, 238–249 (2016).
Assuma, R., Oates, T., Cochran, D., Amar, S. & Graves, D. T. IL-1 and TNF antagonists inhibit the inflammatory response and bone loss in experimental periodontitis. J. Immunol. 160, 403–409 (1998).
Hasturk, H., Kantarci, A. & Van Dyke, T. E. Paradigm shift in the pharmacological management of periodontal diseases. Front. Oral Biol. 15, 160–176 (2012).
Moutsopoulos, N. M. et al. Interleukin-12 and interleukin-23 blockade in leukocyte adhesion deficiency type 1. N. Engl. J. Med. 376, 1141–1146 (2017).
Chen, X. et al. Advanced biomaterials and their potential applications in the treatment of periodontal disease. Crit. Rev. Biotechnol. 36, 760–775 (2016).
Goyal, G., Garg, T., Rath, G. & Goyal, A. K. Current nanotechnological strategies for an effective delivery of drugs in treatment of periodontal disease. Crit. Rev. Ther. Drug Carrier Syst. 31, 89–119 (2014).
Morris, J. J., Lenski, R. E. & Zinser, E. R. The black queen hypothesis: evolution of dependencies through adaptive gene loss. mBio 3, e00036–12 (2012).
Mastellos, D. C., Ricklin, D., Hajishengallis, E., Hajishengallis, G. & Lambris, J. D. Complement therapeutics in inflammatory diseases: promising drug candidates for C3-targeted intervention. Mol. Oral Microbiol. 31, 3–17 (2016).
Van Dyke, T. E. Pro-resolving mediators in the regulation of periodontal disease. Mol. Aspects Med. 58, 21–36 (2017).
Gatej, S., Gully, N., Gibson, R. & Bartold, P. M. Probiotics and periodontitis – a literature review. J. Int. Acad. Periodontol. 19, 42–50 (2017).
Rosier, B. T., Marsh, P. D. & Mira, A. Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J. Dent. Res. 97, 371–380 (2017).
Garza, D. R., van Verk, M. C., Huynen, M. A. & Dutilh, B. E. Towards predicting the environmental metabolome from metagenomics with a mechanistic model. Nat. Microbiol. 3, 456–460 (2018).
Surana, N. K. & Kasper, D. L. Moving beyond microbiome-wide associations to causal microbe identification. Nature 552, 244–247 (2017).
Brown, S. A. & Whiteley, M. A novel exclusion mechanism for carbon resource partitioning in Aggregatibacter actinomycetemcomitans. J. Bacteriol. 189, 6407–6414 (2007).
Brown, S. A. & Whiteley, M. Characterization of the L-lactate dehydrogenase from Aggregatibacter actinomycetemcomitans. PLOS ONE 4, e7864 (2009).
Ramsey, M. M., Rumbaugh, K. P. & Whiteley, M. Metabolite cross-feeding enhances virulence in a model polymicrobial infection. PLOS Pathog. 7, e1002012 (2011).
Stacy, A., Fleming, D., Lamont, R. J., Rumbaugh, K. P. & Whiteley, M. A. Commensal bacterium promotes virulence of an opportunistic pathogen via cross-respiration. mBio 7, e00782–16 (2016).
Stacy, A., Abraham, N., Jorth, P. & Whiteley, M. Microbial community composition impacts pathogen iron availability during polymicrobial infection. PLOS Pathog. 12, e1006084 (2016).
Chawla, A. et al. Community signalling between Streptococcus gordonii and Porphyromonas gingivalis is controlled by the transcriptional regulator CdhR. Mol. Microbiol. 78, 1510–1522 (2010).
Maeda, K. et al. A Porphyromonas gingivalis tyrosine phosphatase is a multifunctional regulator of virulence attributes. Mol. Microbiol. 69, 1153–1164 (2008).
Burns, E., Eliyahu, T., Uematsu, S., Akira, S. & Nussbaum, G. TLR2-dependent inflammatory response to Porphyromonas gingivalis is MyD88 independent, whereas MyD88 is required to clear infection. J. Immunol. 184, 1455–1462 (2010).
Brzezinska, A. A., Johnson, J. L., Munafo, D. B., Ellis, B. A. & Catz, S. D. Signalling mechanisms for Toll-like receptor-activated neutrophil exocytosis: key roles for interleukin-1-receptor-associated kinase-4 and phosphatidylinositol 3-kinase but not Toll/IL-1 receptor (TIR) domain-containing adaptor inducing IFNβ (TRIF). Immunology 127, 386–397 (2009).
Bainbridge, B. et al. Role of Porphyromonas gingivalis phosphoserine phosphatase enzyme SerB in inflammation, immune response, and induction of alveolar bone resorption in rats. Infect. Immun. 78, 4560–4569 (2010).
Acknowledgements
The work of the authors is supported by US National Institutes of Health grants DE01111, DE012505, DE017921 and DE0130585 (R.J.L.); DE018023, DE025220 and DE025848 (H.K.); and DE015254, DE024153, DE024716, DE026152 and AI068730 (G.H.).
Reviewer information
Nature Reviews Microbiology thanks G. Belibasakis, B. Keijser and other anonymous reviewers for their contributions to the peer review of this work.
Author information
Authors and Affiliations
Contributions
R.J.L., H.K. and G.H. researched the data for the article, substantially contributed to discussion of content, wrote the article and reviewed and edited the manuscript before submission.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Glossary
- Subgingival
-
Relating to the area under the gum margins.
- Gingival crevicular fluid
-
(GCF). A serum exudate that contains immune and inflammatory mediators, along with large numbers of neutrophils recruited along a chemokine gradient.
- Extracellular polymeric substances
-
(EPS). Extracellular biomolecules including exopolysaccharides, fibrous and globular proteins in addition to extracellular enzymes, lipids and nucleic acids.
- Gingivitis
-
Mild and reversible inflammation of the gum often accompanied by bleeding upon toothbrushing. Tissue destruction does not occur. Gingivitis results from an accumulation of the plaque biofilm around the gingival margin and resolves after removal of the plaque.
- Acidogenic
-
Organisms capable of producing acidic metabolites and reducing environmental pH.
- Dysbiosis
-
An imbalanced interaction that can be among bacteria in a community or between the microbiome and the host, and is detrimental to the host. The imbalance can be in the amount and/or the influence of individual microbial species relative to their abundance or influence in health. Alternatively, the imbalance can be caused by a poorly controlled immune response.
- Periodontitis
-
An episodic, slowly progressing inflammatory disease of the periodontal tissues that usually occurs in adults, although aggressive, rapidly progressing forms exist and can occur in adolescents.
- Dental caries
-
A polymicrobial and diet-dependent disease that is characterized by the development of pathogenic biofilms (dental plaque) within which acid production from bacterial metabolism of dietary carbohydrates causes demineralization of the mineralized tooth tissues (enamel, dentin and cementum), eventually leading to the clinical onset of cavitation or tooth decay.
- Aciduric
-
Organisms capable of growth at acidic pH levels that are often toxic to other bacteria.
- Polymicrobial synergy
-
Interactions among organisms that increase microbial fitness in the local environment.
- Dentin
-
Calcified tissue, predominantly hydroxyapatite, forming the bulk of the tooth, which is beneath and is softer (less mineralized with more organic material) than enamel.
- Periodontal diseases
-
A collection of conditions in which poorly controlled inflammatory responses induced by the microbiota cause destruction of the supporting structures of the tooth.
- Red complex
-
The triad of P. gingivalis, T. forsythia and T. denticola — organisms that are often isolated together and were classically considered to be the predominant pathogens in chronic periodontitis.
- Gingival crevice
-
The compartment between the tooth root and the gingival (gum) tissue. The gingival crevice deepens into a periodontal pocket as periodontal disease progresses and tissue is destroyed.
- Citrullinate
-
Post-translational modification of a protein involving deamination of arginine by the enzyme peptidylarginine deiminase PPAD to produce citrulline.
- Cross feeding
-
The utilization of a metabolic by-product of one organism as a nutrient source by another organism.
- Nososymbiocity
-
The potential for a microbial community to contribute to disease; this recognizes the community rather than a single species as the aetiological agent.
- Salivary pellicle
-
A layer of salivary proteins and glycoproteins adsorbed to the enamel surface and to which adhesins of initial colonizers of the oral surface can attach. Pellicle can also contain molecules of microbial origin and those derived from epithelial cells.
- Accessory pathogen
-
Organisms that act synergistically with more pathogenic species (keystone pathogens or pathobionts) to elevate community nososymbiocity. Accessory pathogens can provide an attachment substratum for colonization and metabolic support, and can increase virulence gene expression in other organisms through physical interactions or small-molecule-dependent communication.
- Keystone pathogen
-
Species that exert an influence on their communities that is disproportionate relative to their abundance and therefore form the ‘keystone’ of the community’s structure.
- Homeostatic commensals
-
Species that act to maintain a host–microbiota equilibrium by mitigating the action of more pathogenic species. Mechanisms include reducing the impact of pathogens on host cell signalling pathways or production of metabolites that favour a homeostatic inflammatory response.
- Homeostasis
-
A state of equilibrium or stability in a system that is maintained by adjusting physiological processes to counteract external changes.
- Ecological plaque (biofilm) hypothesis
-
A model encompassing microbiological, biochemical and ecological properties of oral biofilms and their association with disease. The model also accommodates host-derived changes (for example, frequent dietary sugar exposure) that trigger changes in the nososymbiocity of biofilm communities.
- Saccharolytic potential
-
The ability of an organism to metabolize carbohydrates.
- Emergent properties
-
Novel structures, activities, patterns and properties that arise during self-organization into complex systems. In the context of biofilm communities, these include surface adhesion and interbacterial cohesion, spatial organization, physical and social interactions, chemical heterogeneity and increased tolerance to antimicrobials.
- Pathobionts
-
Organisms that are generally benign or commensal within an indigenous community but transition to pathogenic upon the breakdown of host–microbiota homeostasis (for example, as a result of antibiotic treatment, tissue damage, dietary shifts and especially immune deficiencies). These conditions promote pathobiont outgrowth and disrupt the symbiotic microbiota, causing further dysbiosis and inflammation.
- Asaccharolytic
-
A property of organisms incapable of breaking down carbohydrates for energy and thus reliant on the degradation of proteins and the generation of amino acids for metabolic energy and growth.
- Inflammophilic
-
A property of bacteria that thrive on inflammation and utilize inflammatory tissue breakdown products for nutrition.
- Hemin
-
An iron-containing porphyrin compound released from red blood cells; exploited by bacteria to obtain iron for growth.
- Localized chemokine paralysis
-
Precise and targeted suppression of specific chemokines by microbial community participants, superseding the otherwise stimulatory activity of other community inhabitants.
- Black queen hypothesis
-
A theory of reductive evolution to account for co-dependency.
Rights and permissions
About this article
Cite this article
Lamont, R.J., Koo, H. & Hajishengallis, G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol 16, 745–759 (2018). https://doi.org/10.1038/s41579-018-0089-x
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41579-018-0089-x
This article is cited by
-
New insights into nanotherapeutics for periodontitis: a triple concerto of antimicrobial activity, immunomodulation and periodontium regeneration
Journal of Nanobiotechnology (2024)
-
Characterization and application of in situ curcumin/ZNP hydrogels for periodontitis treatment
BMC Oral Health (2024)
-
Dental health and lung cancer risk in the Golestan Cohort Study
BMC Cancer (2024)
-
Effects of Sjogren’s syndrome and high sugar diet on oral microbiome in patients with rampant caries: a clinical study
BMC Oral Health (2024)
-
The role of oral microbiota in the development of oral mucositis in pediatric oncology patients treated with antineoplastic drugs: a systematic review
BMC Oral Health (2024)