Abstract
Oral diseases (e.g., dental caries, periodontitis) are developed when the healthy oral microbiome is imbalanced allowing the increase of pathobiont strains. Common practice to prevent or treat such diseases is the use of antiseptics, like chlorhexidine. However, the impact of these antiseptics on the composition and metabolic activity of the oral microbiome is poorly addressed. Using two types of oral biofilms—a 14-species community (more controllable) and human tongue microbiota (more representative)—the impact of short-term chlorhexidine exposure was explored in-depth. In both models, oral biofilms treated with chlorhexidine exhibited a pattern of inactivation (>3 log units) and fast regrowth to the initial bacterial concentrations. Moreover, the chlorhexidine treatment induced profound shifts in microbiota composition and metabolic activity. In some cases, disease associated traits were increased (such as higher abundance of pathobiont strains or shift in high lactate production). Our results highlight the need for alternative treatments that selectively target the disease-associated bacteria in the biofilm without targeting the commensal microorganisms.
Similar content being viewed by others
Introduction
The oral microbiome is one of the most diverse microbial communities that inhabit the human body1. More than 700 bacterial taxa have been identified to date which inhabit different niches in the oral cavity by forming biofilms on surfaces such as the teeth, the gingiva or the tongue (www.homd.org). The oral microbiome is in continuous interaction with environmental factors and its host. Under homeostatic conditions, the oral microbiome is stable and in symbiosis with its host2,3. However environmental perturbations can lead to a shift into dysbiotic biofilms which can be a causative factor of oral diseases, such as caries and periodontitis4,5.
Control of the oral microbiome, prevention and treatment of oral diseases is often achieved with the help of antimicrobials, such as antibiotics and antiseptics. The use of antimicrobials aims at decreasing the total microbial load to tackle the disease. One of the most common antiseptics used in oral health care is chlorhexidine (CHX), a bactericidal agent. CHX has a broad spectrum efficacy and acts by interfering with the cytoplasmic or inner bacterial wall once it has successfully crossed the outside membrane6. CHX is commonly added in treatment products at a concentration of 0.12 or 0.2%. Both concentrations are well above the minimum inhibitory concentration (MIC) of tested oral strains7. Yet, such MIC tests expose the microorganisms to a constant concentration of the biocide. Although this approach is a good proxy for systemic treatment, it is not representative of a topical treatment such as a mouth rinse application. In reality the contact time between the bacteria and the antiseptic is between 60 and 90 s during oral treatment. Moreover in the case of oral diseases, the target is a polymicrobial biofilm and bacteria in biofilms exhibit increased tolerance to antimicrobials compared to planktonic bacteria7. Finally, the biofilm architecture can greatly influence the outcome of the treatment. The outside layers of the biofilm are more susceptible to the antiseptic compared to the inner layers8.
All the above highlight the need for adequate and representative models to study oral biofilms and their response to current or future treatments. These models need to capture the complexity of the biofilm communities but at the same time need to offer controllability and reproducibility. In the field of oral microbiology, synthetic communities consisting of two to six key oral strains have been extensively used to study the interactions between oral microorganisms and their response to external stimuli9,10,11,12,13,14. These simpler synthetic communities allow for a well-controlled system with known players. Their inter-species interactions can be more easily studied and modeled15. Furthermore, species concentrations can be determined accurately. On the opposite side of the experimental spectrum saliva or plaque samples have been used to grow poly-microbial biofilms in vitro16,17. These more complex communities better capture the diversity of an in vivo oral biofilm and the interactions between the oral microorganisms. It is a step closer to a more realistic model. Yet, the increased complexity of these systems leads to reduced controllability and require more elaborate methods to track the community response and dynamics. As a result, studies assessing the effect of antiseptics on in vitro biofilms from saliva or plaque origin have primarily focused on microscopic techniques8,18. Such an approach studies the response of a biofilm as one unit and does not examine the community composition. Only few studies have tried to resolve the above by using amplicon sequencing to track community shifts19. However, the outcome of the treatment might be greatly influenced by the composition of the surviving community, making it very important to identify it while studying the effect of the antimicrobial stress on the polymicrobial biofilm.
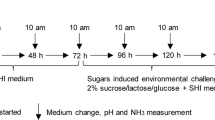
The aim of the present study was to evaluate the impact of antiseptic treatment on in vitro oral biofilms. Previous studies have shown that mouthwash rinsing, even when applied consecutively for many days, had only a temporal effect20. We hypothesized that the treatment would not only affect the living cells concentration but also the composition and metabolic activity of the surviving community, and that consecutive exposures could enhance this phenomenon. We used two different microbial communities to mimic the polymicrobial oral biofilm: (i) a 14-strain biofilm and (ii) a tongue swab derived microbiota biofilm. The biofilms were exposed for a short (5 min) period to 0.12% CHX every 24 h to simulate the oral care procedure and we dynamically monitored the cell viability, community composition, and metabolic activity.
Results
A dynamic in vitro model was used to mimic oral biofilms and study their response to antiseptic stress. A 14-species synthetic community or a tongue swab from four subjects was used as an inoculum and the response to CHX was evaluated in terms of microbial survival and regrowth, community composition, and metabolic activity.
CHX leads to initial drop in biofilm bacterial cell concentration followed by quick recovery
To test the effect of the antiseptic treatment on cell viability, in vitro oral biofilms were treated for 5 min with 0.12% CHX during three consecutive days. Microbial survival was determined by means of flow cytometry and SGPI (viability) staining. The cells were separated in three clusters based on their flow cytometric profile: intact, damaged, and dead.
The initial concentration of intact cells in the 14-species biofilms was ~107 cells/cm2. Control biofilms that were not exposed to CHX exhibited a constant growth over time to a final concentration of 1011 intact cells/cm2 at the end of the experiment. In contrast, the first short exposure to CHX resulted in a large decrease of intact cells with three log units (to 104 cells/cm2), yet without completely inactivating the biofilm (Fig. 1a). As expected, an increase in damaged and dead cells was observed after the treatment. The intact cell concentration remained stable for the following 24 and 48 h despite the two additional CHX treatments. At 72 h, after three consecutive days of short CHX exposures, however, the bacterial concentration sharply rose to 107 cells/cm2 (similar to the concentration at 0 h) (Fig. 1a). The more complex tongue-swab derived biofilms displayed a similar pattern of inactivation and regrowth (Fig. 1b). However their regrowth response was even faster and took already place after 24 h. More in detail, the initial intact cell concentration was 106 cells/cm2 and dropped to 104 cells/cm2 immediately after exposure to CHX. Yet, the following 24 h of incubation displayed a 2 log regrowth. The following CHX exposures at 48 and 72 h showed similar patterns: a drop in intact cell concentration immediately after CHX treatment, followed by a rapid regrowth in the subsequent incubation period, eventually bringing back the intact cell concentration to the original value of 107 cells/cm2. This pattern was consistent for all incubations with human derived oral biofilms. As expected, the dynamics of damaged and dead cell concentrations were the opposite of those from the intact cells.
Concentrations of live, damaged, and dead cells for a. 14-species biofilms. Points are the average of four replicates and error bars represent the standard deviation. b For tongue deriving microbiota biofilm from four individuals (donors). The red line is the concentration of CHX treated biofilms, purple/blue for the non-treated control biofilms. The vertical red lines represent the points of treatment.
Biofilm composition shift after CHX treatment
We hypothesized that the antiseptic treatment alters the microbial composition of the biofilm, as it is already known that susceptibility to CHX is strain dependent. To determine how the composition changed over time, the abundances of the different taxa were measured either by means of qPCR (for the 14 strain-biofilm) or by 16S rRNA gene amplicon sequencing (for the tongue microbiota derived biofilms).
Non-treated 14 strains biofilms were highly dominated by Veillonella parvula (99% relative abundance, Fig. 2). In contrast, CHX exposure had a big impact on community composition as V. parvula relative abundance rapidly decreased to >5%, while Streptococcus gordonii showed a big increase in abundance reaching 94% (±0.08) of the total biofilm at 72 h. The six pathobionts (P. gingivalis, P. intermedia, A. actinomycontaminants, F. nucleatum, S. mutans, and S. sobrinus) were present at 3–4 log concentrations lower in the biofilms compared to the more dominant strains (A. actinomycontaminants <104 cells/cm2 while P. gingivalis, P. intermedia, F. nucleatum, S. mutans, and S. sobrinus were <103 cells/cm2). However, these concentrations were unaffected by CHX and thus remained stable over the course of the treatment.
16S rRNA gene amplicon sequence analysis revealed the initial composition of the tongue-swab derived biofilms to be donor-dependent (Fig. 3). This inter-individual difference became less clear under control conditions as β-diversity of mature biofilms dropped over time with samples clustering together in non-metric multidimensional scaling (NMDS) ordination plots based on Bray–Curtis dissimilarities (Fig. 3). In sharp contrast, tongue-swab derived biofilms exposed to CHX were diverging from each other over time: a clear donor-dependent effect was observed.
At baseline all tongue microbiota derived biofilms were dominated by Veillonella and Streptococcus, 84–97% were classified as Veillonellaceae and Streptococcaceae (Fig. 4). Over the course of the treatment donor-dependent changes were observed in the most abundant taxa. Donor 1-derived biofilms exhibited a small decrease in relative abundances of Veillonella. Donor 2-biofilm displayed a large shift in the composition after the first CHX treatment with an increase in the relative abundance of the genus Granulicatella after the first two treatments. Relative abundances of Fusobacterium, Haemophilus and Solobacterium increased during the course of treatment in donor 3-biofilm. Finally, donor 4-biofilm exhibited the most profound community shift. The composition shifted towards a Fusobacterium monodominance with this genus accounting for more than 90% of the total community.
Metabolic activity shifted after CHX treatment reflecting the community shift
To investigate the effect of antiseptic stress on the metabolic activity of the community, the ability to produce or consume organic acids was evaluated. The concentration of the lactate, acetate, propionate, formate and butyrate on surrounding medium was measured every 24 h.
Non-treated 14-strains biofilms produced propionate (18.96 ± 0.6 mM/day), acetate (13.49 ± 0.19 mM/day) and formate (2.83 ± 0.68 mM/day) after the first 24 h (Fig. 5). The relative production of organic acids remained constant over time in maturing (non-treated) biofilms. In contrast, total organic acid production by CHX-treated biofilms greatly decreased to 4.76 ± 0.36 mM/day after the first exposure to antiseptics, compared to non-treated biofilms, which produced 43.15 ± 0.33 mM/day. After the initial decrease, total organic acid production recovered but with a different composition, shifting to a high production of lactate (23.1 ± 1.00 mM).
Non-treated in vitro tongue microbiota biofilms produced acetate (25.35 ± 1.34 mM/day), propionate (10.24 ± 1.00 mM/day) and lower concentrations of formate (3.01 ± 2.4 mM/day) and butyrate (2.96 ± 2.87 mM/day) (total 49.84 ± 3.2 mM/day). Total production increased over time (69.06 ± 11.38 mM/day) as well as the relative concentration of butyrate (accounting for 14–23%). Small differences were observed between donors. On the other hand, CHX-treated samples produced much lower concentrations of organic acids (8.03 ± 5.62 mM/day). Notably the donor effect was again obvious. For donor 1 and 2 relative production did not change with acetate and propionate being the organic acids produced at higher concentrations. However, in donor 4-biofilm a higher production of butyrate (+1.5 mM/day compared to the other donors) was observed. These results correspond with the community composition shift and the high relative abundances of Fusobacterium in the same biofilm.
Discussion
Oral biofilms are polymicrobial communities with a vital role in oral and systemic health. Oral care commonly includes the use of wide spectrum antimicrobials (i.e., chlorhexidine). However, these non-targeted treatments can have a big impact on the innate commensal microbial community. In fact, previous studies suggest that antiseptic stress affects oral biofilm composition and metabolic activity19. Meanwhile the long-term effect and dynamic response from the oral biofilms to antiseptics is poorly understood. In this study we investigated the response of in vitro oral biofilms to consecutive treatments with a wide spectrum antiseptic, i.e., CHX. We evaluated the impact from short daily CHX exposures on microbiota composition and activity with two model systems for oral biofilms: (i) a biofilm derived from a synthetic community composed of 14 commensal and pathobiont oral strains, being more controllable because of its defined composition and (ii) a set of biofilms derived from tongue swab microbiota from healthy individuals, being a closer proxy to the complexity of the in vivo microbiota. Although moderate model-dependent differences in the biofilm response to CHX exposure were noted, the overall trend of CHX affecting community composition and functionality was consistent in both model systems.
Simulating a mouthwash procedure, short daily exposures to 0.12% CHX exhibited a repeated pattern of inactivation and rapid regrowth in both biofilm types. Although chlorhexidine exhibits a high substantivity which means that the antimicrobial action will continue longer than the actual rinse21, our findings show CHX to only have a temporal effect on oral bacterial biofilms which is supported by both in vivo and in vitro previous studies18,22. These results indicate that oral antiseptics will fall short of keeping microbial numbers under control and are thus ineffective in maintaining oral hygiene. Moreover, as broad-spectrum antiseptics such as CHX also affect the endogenous oral microbiota, there is an increased risk that microbial dysbiosis in the oral cavity will occur resulting in the development of oral diseases. Indeed, our results confirm that the community composition from both synthetic as natural oral biofilms are drastically altered upon repeated CHX exposure. The 14 species-biofilms displayed profound shifts from a V. parvula dominated community (99%) to a Streptococcus dominated community, more specifically Streptococcus gordonii (98%) upon CHX treatment. The higher tolerance of S. gordonii to CHX, as exhibited by a CHX susceptibility test (MIC), may explain this result (Supplementary Table 2). Veillonella species are common colonizers of the oral cavity and are able to produce propionate from lactate23, which is typically produced by Streptococcus species. It was therefore not a surprise that the observed phylogenetic shifts coincided with drastic changes in metabolite profiles. Untreated biofilms mainly produced propionate (23.3 mM) while CHX exposed biofilms mainly produced lactate (23.1 mM ± 1) (Fig. 5). Streptococcus gordonii, the most abundant strain in the CHX treated 14-strain biofilms, is considered a primary colonizer of the dental surface24 and produces l-lactate as primary metabolite. While it is less aciduric than mutans streptococci25, it succeeded in dominating the synthetic oral biofilm despite the significant decrease in pH from 7 to pH 5.8 during the incubation experiment. High concentrations of lactate in combination with a low pH (the pKa of lactic acid/lactate being 3.86) are important determinants of tooth demineralization and tooth decay and increase the risk for dental caries. At the same time, the concentrations of the pathobiont strains were not reduced (with the exception of S. mutans). This outcome cannot be explained by the individual tolerance to chlorhexidine as most strains exhibit similar or lower MIC than other strains which relative abundance decreased drastically (e.g., V. parvula). The higher survival rates of the pathobiont strains could be explained by the biogeography of the oral biofilm, where these strains normally inhabit the inner layers of the biofilm and they are thus more protected26.
The bacterial community of the tongue microbiota biofilms was also affected by CHX treatments. The most pronounced shift was observed in the biofilms derived from donor 4 where there was a continuous increase of the relative abundance of the genus Fusobacterium over the course of the daily CHX treatments. The increased dominance of Fusobacterium is significant because it is considered a bridge organism between early and late colonizers in oral biofilms facilitating biofilm maturation and attachment of pathobionts. This genus is prevalent in periodontal plaque samples27,28 and thus linked to periodontal disease. The observed higher production of butyrate by the CHX exposed natural biofilm is also indicative of higher Fusobacterium dominance23,29. Butyrate is a bacterial metabolic by-product with detrimental effects for oral health, triggering inflammation in gingival fibroblasts30 and disrupting the gingival epithelial barrier31. With respect to the other in vivo derived biofilms, we found the antiseptic treatment to impact the biofilms in a dynamic and donor-dependent manner, with composition and metabolic activity shifting after every treatment. Despite the fact that non-treated mature biofilms clustered together according to β-diversity, the community composition of CHX treated biofilms was clearly dependent on initial inoculum. Our results confirm previous observations where inter-personal microbiome variability was described as one of the main drivers in response to oral treatment32,33.
Overall our findings made evident that wide spectrum antimicrobials cannot guarantee a shift to a healthy state. On the contrary they can further perturb the commensal microbiome. These findings are in accordance with recent in vivo study that shows that CHX treatment although kept microbial load in lower numbers than no treatment, did so by unselective targeting of the oral microbiome which resulted in higher relative abundances of several periodontitis related taxa (i.e., Fusobacterium)34. Moreover antiseptics have been already shown to increase pathogenic characteristics35, resistance to antiseptics36, and cytotoxicity to host cells37. For this reason there is a clear need for alternative approaches that do not indiscriminately target the oral microbiome but would specifically and selectively target pathogenic strains38 (or their virulence—virulence therapy) and promote or re-establish a healthy microbiome (pro-biotics and pre-biotics)39,40,41.
To conclude, we state that antiseptics are inadequate as sole treatment of oral biofilms. We observed that short treatment with 0.12% CHX, a concentration commonly used in oral care products, only temporarily decreased the viable cell concentration. Furthermore, our results suggest that initial microbiome composition highly influenced the outcome of the antiseptic treatment with disease associated characteristics increasing after treatment in certain cases. Further in vivo experiments are required to elucidate the clinical relevance of this study and the possible implications of the extensive use of antimicrobials in oral care.
Materials and methods
Strains and culture
The synthetic community was assembled by 14 oral bacterial strains, eight commensal (Streptococcus sanguinis LMG14657, Streptococcus salivarius TOVE-R, Streptococcus gordonii ATCC 49818, Streptococcus mitis DSM 12643, Streptococcus oralis (clinical isolate), Actinomyces naeslundii ATCC 51655, Actinomyces viscosus DSM 43327 and Veillonella parvula DSM 2007) and six pathobionts (Porphyromonas gingivalis ATCC 33277, Fusobacterium nucleatum ATCC10953, Aggregatibacter actinomycetemcomitans ATCC 43718, Prevotella intermedia ATCC 25611, Streptococcus mutans ATCC 25175 and Streptococcus sobrinus ATCC 33478). The strains were maintained on blood agar No2 (Oxoid, Hampshire,UK) supplemented with hemin (5 mg/mL) (Sigma Aldrich, Belgium), menadione (1 mg/mL) (Sigma Aldrich, Belgium) and 5% sterile horse blood or cultured in liquid medium in Brain Hearth Infusion (BHI) (Roche, Belgium) broth under anaerobic (80% N2, 10% H2, and 10% CO2) conditions.
BHI medium was used for the assembled synthetic community. This medium is enriched with 2.5 g/L Mucin from porcine stomach type III (Sigma, Diegem, Belgium), 1.0 g/L Yeast extract (Oxoid, Hampshire, UK), 0.1 g/L cysteine (Merck—Calbiochem), 2.0 g/L sodium bicarbonate (Sigma Aldrich, Belgium), 0.25% glutamic acid (Merck—Calbiochem), 5.0 mg/L hemin (Sigma Aldrich, Belgium), 1.0 mg/L menadione (Sigma Aldrich, Belgium).
Minimum inhibitory concentration
The MIC of CHX for each individual strain was evaluated by absorbance. More specifically, 107 cells/mL of each strain was inoculated in serial dilutions of CHX and incubated for 24 h under anaerobic conditions. Following OD600 was measured by microtiter plate reader (Tecan Infinite M200 Pro; Tecan UK, Reading, UK).
Tongue microbiome sampling
The tongue microbiome samples originated from four individuals. The individuals were in good systemic health and had not received antibiotics over the previous 3 months. The donors were asked to refrain from food intake or tongue/tooth brushing for at least 2 h before collection of the tongue biofilm.
Tongue biofilm was sampled by scraping the tongue surface with a disposable tongue scraper (Jordan, Norway) three times from back to front of the tongue dorsum. The scrapped biofilm was suspended in 5 mL of reducing phosphate buffer (8.8 g/L K2HPO4, 6.8 g/L KH2PO4 and 1 g/L C2H3O2SNa (Sigma Aldrich, Belgium)) and homogenized by pipetting keeping individual samples separate. One milliliter of each sample was used to inoculate a separate anaerobic penicillin bottle with 15 mL BHI 2 medium. Samples were allowed to separately grow in planktonic conditions for 48 h before used further to grow a biofilm.
Biofilm model
After the 14 strains had grown individually in BHI broth as described above, the synthetic community was assembled by adding equal concentrations of each strain in BHI 2 medium. Then they were allowed to grow for 48 h under anaerobic (80% N2,10% H2, and 10% CO2) conditions. Subsequently inoculum from the planktonic bioreactor was used to grow biofilms on hydroxyapatite (HA) disks (0.5 inch diameter × 0.04–0.06 inch thick, VWR, Belgium) that were placed in an active attachment biofilm model42. Sample of the bioreactor was diluted 1:2 v/v in fresh BHI 2 medium. Two milliliter of the new culture containing the community were added in each well of a 24-well plate and incubated for 24 h at 37 °C in anaerobic conditions and shaking at 170 rpm. Twenty-fours hours-biofilms were used for the following experiments.
The same procedure was followed with the donor samples that had been previously grown in BHI 2 for 48 h.
Treatment
The biofilms were treated with CHX by placing the HA disks for 5 min in a new 24-well plate with 0.12% CHX (Chlorohexidine digluconate, Sigma Aldrich, Belgium). Non-treated biofilms underwent the same procedure in sterile PBS (Phosphate Buffer Saline, Sigma Aldrich, Belgium). They were subsequently washed twice with PBS and finally placed back in a new 24-well plate with fresh BHI 2 medium. The biofilms were treated every 24 h and samples were collected before and after treatment. Biofilms were disrupted by Trypsin-EDTA 0.05% at 37 °C, 350 rpm for 45 min and then further placed in a sonication bath (37 kHz sonication frequency) for 32 min to disrupt the aggregates.
Flow cytometry
Samples of the disrupted biofilm were diluted in sterile PBS and stained with the nucleic acid stain SYBR® Green I and Propidium Iodide that stains permeabilized cells43. SYBR Green I (10,000× concentrate in DMSO, Invitrogen) was diluted 100 times in 0.22 µm-filtered DMSO (IC Millex, Merck, USA) and Propidium Iodide (20 mM in dimethyl sulfoxide (DMSO), Invitrogen, USA) was diluted 50 times. Samples were stained with 10 µL/mL staining solution. Next, they were incubated in 37 °C for 13 min. All samples were measured with a benchtop Accuri C6+ cytometer (BD Biosciences, Belgium). The stability of the instrument was controlled daily using CS&T RUO beads (BD Biosciences, Belgium) and checking for each measurement the stability of FL1 over time. The blue laser (488 nm)was used for the excitation of the stains. The filters for the (fixed gain) photomultiplier detectors used during the measurements were 533 nm with a bandpass of 30 nm for the green fluorescence (FL-1) and 670 nm longpass filter for the red fluorescence (FL-3). The threshold was set on the 533/30 nm (FL-1) detector at the arbitrary unit of 500. FlTC-A ~ PerCP-A density plots were used to draw gates for intact, damaged and dead cells according to Supplementary Fig. 1.
DNA extraction and vitality qPCR of the 14-strain biofilms
The disrupted biofilms were treated with PMAxx Dye (Biotium, USA) before further manipulation. DNA extraction was performed by means of bead beating with a PowerLyzer instrument (Qiagen, Venlo, Netherlands) and phenol/chloroform extraction. The surviving bacteria were quantified by vitality qPCR using specific primers44,45. The qPCR measurements were performed with a CFX96 Real-Time System (Bio-Rad, Temse, Belgium). In short, Taqman 5′ nuclease assay PCR method was used for detection and quantification of every one of the 14 strains using strain specific primers and probes (Supplementary Table 1). Quantification was based on a plasmid standard curve.
DNA extraction and amplicon sequencing of multispecies biofilms
DNA from the multi-species biofilms was extracted using the ZymoBIOMICS DNA Microprep Kit (Zymo Research, USA) according to the manufacturer's instructions. 10µl genomic DNA extract was send out to LGC genomics GmbH (Berlin, Germany) for library preparation and sequencing on an Illumina Miseq platform with v3 chemistry with the primers 341F (5′-CCT ACG GGN GGC WGC AG -3′) and 785Rmod (5′-GAC TAC HVG GGT ATC TAA KCC-3′)46.
The average number of raw reads per sample was 45712. Read assembly and cleanup was largely derived from the MiSeq SOP described by the Schloss lab. In brief, mothur (v.1.40.3) was used to assemble reads into contigs, perform alignment-based quality filtering (alignment to the mothur-reconstructed SILVA SEED alignment, v. 123), remove chimeras, assign taxonomy using a naïve Bayesian classifier47 and SILVA NR v132 and cluster contigs into OTUs at 97% sequence similarity. All sequences that were classified as Eukaryota, Archaea, Chloroplasts and Mitochondria were removed. Also, if sequences could not be classified at all (even at (super)Kingdom level) they were removed. After the above-mentioned filtering, the average reads per sample were 27940. For each OTU representative sequences were picked as the most abundant sequence within that OTU.
Metabolic activity
Organic acids in the samples were measured with 761 Compact Ion Chromatograph (Metrohm, Switzerland) with a Metrosep Organic acids 250/7.8 column and a Metrosep Organic acids Guard/4.6 guard column. The eluent consists of 1 mM H2SO4 at a flow rate of 0.8 mL/min. The production of organic acids was calculated as the concentration detected in the liquid that the biofilm was grown normalized for the organic acid concentrations detected in sterile BHI 2 medium.
Data analysis/statistics
All statistical analysis was performed in R (v3.6.0). The OTU contingency table was imported in R. OTUs with no more than one read in every sample (singletons) were removed48. The average number of reads per sample after removing singletons was 27866, while the total number of OTUs was 2350. The graphs representing the 20 most relative abundant genera were generated using the phyloseq package49 in R (v3.6.0). The taxonomic β-diversity was calculated based on Bray-Curtis dissimilarity index, using the ordinate function of phyloseq package, and displayed in a non-metric multidimensional scaling (nMDS) plot.
Ethics statement
The sampling of human tongue biofilm was approved by the the Medical Ethical Committee of Ghent University with reference number B670201629302. All participants gave their written informed consent prior to their inclusion in the study.
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
Sequences are available on the NCBI Sequence Read Archive (SRA) under accession number PRJNA554992.
References
Consortium, T. H. M. P. Structure, function and diversity of the healthy human microbiome. Nature 486, 207–214 (2012).
Hezel, M. P. & Weitzberg, E. The oral microbiome and nitric oxide homoeostasis. Oral Dis. 21, 7–16 (2015).
Rosier, B. T., Marsh, P. D. & Mira, A. Resilience of the oral microbiota in health: mechanisms that prevent dysbiosis. J. Dent. Res. 97, 371–380 (2017).
Bergstrom, J. Smoking rate and periodontal disease prevalence: 40-year trends in Sweden 1970–2010. J. Clin. Periodontol. 41, 952–957 (2014).
Moynihan, P. J. & Kelly, S. A. M. Effect on caries of restricting sugars intake: systematic review to Inform WHO guidelines. J. Dent. Res. 93, 8–18 (2014).
Donnell, G. M. C. Antiseptics and disinfectants: activity, action, and resistance. Clin. Microbiol. Rev. 12, 147–179 (1999).
Mcbain, A. J. et al. Effects of a chlorhexidine gluconate-containing mouthwash on the vitality and antimicrobial susceptibility of in vitro oral bacterial ecosystems. Appl. Environ. Microbiol. 69, 4770–4776 (2003).
Zaura-Arite, E., van Marle, J. & ten Cate, J. M. Conofocal microscopy study of undisturbed and chlorhexidine-treated dental biofilm. J. Dent. Res. 80, 1436–1440 (2001).
Thurnheer, T., Gmu, R. & Guggenheim, B. Multiplex FISH analysis of a six-species bacterial biofilm. J. Microbiol. Methods 56, 37–47 (2004).
Guggenheimrl, B., Giertsen, E., Schupbachl, P. & Shapirol, S. Validation of an in vitro biofilm model of supragingival plaque. J. Dent. Res. 80, 363–370 (2001).
Pratten, J., Barnett, P. & Wilson, M. Composition and susceptibility to chlorhexidine of multispecies biofilms of oral bacteria. Appl. Environ. Microbiol. 64, 3515–3519 (1998).
Seneviratne, C. J. et al. Nanoparticle-encapsulated chlorhexidine against oral bacterial biofilms. PLoS ONE 9, e103234 (2014).
Peyyala, R., Kirakodu, S. S., Ebersole, J. L. & Novak, K. F. Novel model for multispecies biofilms that uses rigid gas-permeable lenses. Appl. Environ. Microbiol. 77, 3413–3421 (2011).
Kommerein, N. et al. An oral multispecies biofilm model for high content screening applications. PLoS ONE 12, e0173973 (2017).
Chan, H. S. J., Simons, M. N. & Maranas, C. D. SteadyCom: predicting microbial abundances while ensuring community stability. PLoS Comput. Biol. 13, e1005539 (2017).
Fernandez y Mostajo, M. et al. A reproducible microcosm biofilm model of subgingival microbial communities. J. Periodontal Res. 52, 1021–1031 (2017).
Nance, W. C. et al. A high-throughput microfluidic dental plaque biofilm system to visualize and quantify the effect of antimicrobials. J. Antimicrob. Chemother. 68, 2550–2560 (2013).
Shen, Y. et al. Experimental and theoretical investigation of multispecies oral biofilm resistance to chlorhexidine treatment. Sci. Rep. 6, 27537 (2016).
Fernandez y Mostajo, M., Exterkate, R. A. M., Buijs, M. J., Crielaard, W. & Zaura, E. Effect of mouthwashes on the composition and metabolic activity of oral biofilms grown in vitro. Clin. Oral Investig. https://doi.org/10.1007/s00784-016-1876-2 (2016).
Gazaniga, L., Ribeiro, M., Hashizume, L. N. & Maltz, M. The effect of different formulations of chlorhexidine in reducing levels of mutans streptococci in the oral cavity: a systematic review of the literature. J. Dent. 35, 359–370 (2007).
Quintas, V., Prada-Lopez, I., Donos, N., Suarez-Quintanilla, D. & Toma, I. In situ neutralisation of the antibacterial effect of 0.2% chlorhexidine on salivary microbiota: quantification of substantivity. Arch. Oral. Biol. 60, 1109–1116 (2015).
García-Caballero, L. et al. Chlorhexidine substantivity on salivary flora and plaque-like biofilm: an in situ model. PLoS ONE 8, 1–10 (2013).
Takahashi, N. Oral microbiome metabolism: From “ Who Are They?” to “ What Are They Doing?”. Crit. Rev. Oral. Biol. Med. 94, 1628–1637 (2015).
Diaz, P. I. et al. Molecular characterization of subject-specific oral microflora during initial colonization of enamel. Appl. Environ. Microbiol. 72, 2837–2848 (2006).
de Soet, J. J., Nyvad, B. & Kilian, M. Strain-related acid production by oral. Caries Res. 34, 486–490 (2000).
Mark, J. L., Rossetti, B. J., Rieken, C. W., Dewhirst, F. E. & Borisy, G. G. Biogeography of a human oral microbiome at the micron scale. PNAS 791–800 (2016). https://doi.org/10.1073/pnas.1522149113.
Li, Y. et al. Phylogenetic and functional gene structure shifts of the oral microbiomes in periodontitis patients. ISME J. 8, 1879–1891 (2014).
Rocas, I. N. & Siqueira, J. F. Root canal microbiota of teeth with chronic apical periodontitis. J. Clin. Microbiol. 46, 3599–3606 (2008).
Jorth, P., Turner, K. H., Gumus, P., Nizam, N. & Buduneli, N. Metatranscriptomics of the human oral microbiome during health and disease. MBio 5, e01012–e01014 (2014).
Chang, M.-C. et al. Butyrate induces reactive oxygen species production and affects cell cycle progression in human gingival fibroblasts. J. Periodontal Res. 48, 66–73 (2013).
Liu, J. et al. Butyrate rather than LPS subverts gingival epithelial homeostasis by downregulation of intercellular junctions and triggering pyroptosis. J. Clin. Periodontol. 46, 894–907 (2019).
Filoche, S. K., Soma, D., Van Bekkum, M. & Sissons, C. H. Plaques from different individuals yield different microbiota responses to oral-antiseptic treatment. FEMS Immunol. Med. Microbiol. 54, 27–36 (2008).
Bizzarro, S. et al. Microbial profiles at baseline and not the use of antibiotics determine the clinical outcome of the treatment of chronic periodontitis. Sci. Rep. 6, 1–13 (2016).
Al-kamel, A. et al. Subgingival microbiome of experimental gingivitis: shifts associated with the use of chlorhexidine and N-acetyl cysteine mouthwashes. J. Oral Microbiol. 11, 1608141 (2019).
Liu, G. et al. Shift in the subgingival microbiome following scaling and root planing in generalized aggressive periodontitis. J. Clin. Periodontol. 45, 440–452 (2018).
Verspecht, T. et al. Development of antiseptic adaptation and cross-adapatation in selected oral pathogens in vitro. Sci. Rep. https://doi.org/10.1038/s41598-019-44822-y (2019).
Müller, H. D., Eick, S., Moritz, A., Lussi, A. & Gruber, R. Cytotoxicity and antimicrobial activity of oral rinses in vitro. Biomed. Res. Int. 2017, 4019723 (2017).
Page, R. C. et al. Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis. Oral Microbiol. Immunol. 22, 162–168 (2007).
Teughels, W. et al. Clinical and microbiological effects of Lactobacillus reuteri probiotics in the treatment of chronic periodontitis: a randomized placebo-controlled study. J. Clin. Periodontol. 40, 1025–1035 (2013).
López-López, A., Camelo-Castillo, A., Ferrer, M. D., Simon-Soro, A. & Mira, A. Health-associated niche inhabitants as oral probiotics: the case of Streptococcus dentisani. Front. Microbiol. 8, 1–12 (2017).
Slomka, V. et al. Nutritional stimulation of commensal oral bacteria suppresses pathogens: the prebiotic concept. J. Clin. Periodontol. 44, 344–352 (2017).
Exterkate, R. A. M., Crielaard, W. & Ten Cate, J. M. Different response to amine fluoride by streptococcus mutans and polymicrobial biofilms in a novel high-throughput active attachment model. Caries Res. 44, 372–379 (2010).
Van Nevel, S., Koetzsch, S., Weilenmann, H., Boon, N. & Hammes, F. Routine bacterial analysis with automated flow cytometry. J. Microbiol. Methods 94, 73–76 (2013).
Loozen, G., Boon, N., Pauwels, M., Quirynen, M. & Teughels, W. Live/dead real-time polymerase chain reaction to assess new therapies against dental plaque-related pathologies. Mol. Oral Microbiol 26, 253–261 (2011).
Herrero, E. R. et al. Dysbiotic biofilms deregulate the periodontal inflammatory response. J. Dent. Res. 97, 547–555 (2018).
Klindworth, A. et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 41, 1–11 (2013).
Wang, Q., Garrity, G. M., Tiedje, J. M., Cole, J. R. & Al, W. E. T. Naıve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267 (2007).
McMurdie, P. J. & Holmes, S. Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput. Biol. 10, e1003531 (2014).
Mcmurdie, P. J. & Holmes, S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Acknowledgements
This work was supported by ‘Fonds voor Wetenschappelijk Onderzoek’—FWO (G0B2719N). We thank Cristina Garcia Timermans, Jasmine Heyse and Josefien Van Landuyt for critically reading the manuscript.
Author information
Authors and Affiliations
Contributions
All authors conceived and designed the study. I.C. performed the experiments and analyzed the data. I.C. wrote the manuscript. All authors reviewed and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chatzigiannidou, I., Teughels, W., Van de Wiele, T. et al. Oral biofilms exposure to chlorhexidine results in altered microbial composition and metabolic profile. npj Biofilms Microbiomes 6, 13 (2020). https://doi.org/10.1038/s41522-020-0124-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41522-020-0124-3
This article is cited by
-
Promising dawn in tumor microenvironment therapy: engineering oral bacteria
International Journal of Oral Science (2024)
-
The efficacy of mouthwashes on oral microorganisms and gingivitis in patients undergoing orthodontic treatment: a systematic review and meta-analysis
BMC Oral Health (2023)
-
Preprocedural mouthwashes for infection control in dentistry—an update
Clinical Oral Investigations (2023)
-
Comparison of the modulatory effects of three structurally similar potential prebiotic substrates on an in vitro multi-species oral biofilm
Scientific Reports (2021)
-
Limited antimicrobial efficacy of oral care antiseptics in microcosm biofilms and phenotypic adaptation of bacteria upon repeated exposure
Clinical Oral Investigations (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.