Abstract
Lung squamous cell carcinoma (LSCC) is a prevalent and progressive subtype of lung cancer. This study aimed to substantiate the regulatory effect of the PAK2/SOX2/DEK axis on the LSCC development. LSCC tissues (n = 83) and adjacent normal tissues were collected and SOX2 expression was determined by qRT-PCR and Western blotting. Correlation between SOX2 expression and the prognosis of LSCC patients was then explored utilizing Kaplan–Meier analysis. Co-immunoprecipitation and glutathione-S-transferase pull-down assays were conducted to validate the binding of SOX2 to DEK. Gain- and loss- of function assays were then performed on LSCC cells, with CCK-8 and Transwell assays applied to detect the malignant behaviors of cells. A mouse xenograft model of LSCC was further established for in vivo validation. The expression levels of SOX2, PAK2 and DEK were up-regulated in LSCC tissues and cells. SOX2 overexpression was correlated with poor prognosis of LSCC patients. Knockdown of SOX2 weakened the viability and the migratory and invasive potential of LSCC cells. Further, PAK2 directly interacted with SOX2. PAK2 overexpression accelerated the malignant phenotypes of LSCC cells through interplay with SOX2. Moreover, SOX2 activated the expression of DEK, and silencing DEK attenuated the malignant behaviors of LSCC cells. In conclusion, PAK2 could bind to the transcription factor SOX2 and thus activate the expression of DEK, thereby driving the malignant phenotypes of LSCC cells both in vivo and in vitro.
Similar content being viewed by others
Introduction
Lung cancer is one of the leading causes of cancer-related mortality worldwide1. Lung cancer is generally divided into two types, small cell lung carcinoma and non-small cell lung carcinoma (NSCLC), and NSCLC can further be classified into large cell lung carcinoma, lung squamous cell carcinoma (LSCC), and lung adenocarcinoma2. Among the three subtypes, LSCC accounts for about 30% of NSCLC and is characterized by poor survival rates and frequent recurrence3. In spite of considerable efforts to develop LSCC treatment, several available therapeutic regimens, including a newly developed targeted therapy, fail to achieve satisfactory therapeutic results, which may be attributed to the complicated genomic patterns in LSCC4. Therefore, the identification of novel targets is urgent for the development of LSCC treatment.
SOX2, a transcription factor that acts as a regulator of pluripotent stem cells, has been identified as a contributor to the development and maintenance of squamous epithelia5. The amplification of SOX2 as well as its oncogenicity has been reported in a spectrum of squamous cell carcinomas (SCCs)6,7. More importantly, previous studies have revealed the involvement of SOX2 in LSCC8. However, the molecular mechanism by which SOX2 participates in LSCC progression remains to be established. Our bioinformatics analysis initially identified p21-activated kinase 2 (PAK2) as a SOX2-interacting protein, and, notably, the interaction between SOX2 and PAK2 was revealed in a previous systematic analysis9. Interestingly, a recent case has correlated PAK overexpression with oral SCC10, whereas the potential role of PAK2 in LSCC remains unclear. This study therefore aimed to explore the potential effect of PAK2 on LSCC based on its interaction with SOX2.
Apart from the interaction between SOX2 and PAK2, it has also been suggested that SOX2-mediated up-regulation of defective kernel (DEK) contributed to the regulatory effect of SOX2 in LSCC5. DEK, a typical oncogene, is involved in the progression of various cancers11. Moreover the expression of DEK was up-regulated in NSCLC, and DEK overexpression was involved in the pathogenesis of NSCLC, stimulating the proliferation and invasion of NSCLC cells12,13,14. In this study, we hypothesize that PAK2 induces the occurrence and development of LSCC by positively interacting with SOX2 and up-regulating the expression of DEK.
Methods and materials
Bioinformatics analysis
LSCC-related genes were retrieved, with the key word “lung squamous cell carcinoma”, from GeneCards (Relevance score: top 300 genes), DisGeNET (screen criteria: score ≥ 0.1) and Phenolyzer databases, respectively, which were then intersected using the jvenn tool to identify candidate genes. Protein-protein interaction (PPI) analysis was conducted through a bioinformatics tool STRING (confidence 0.4) to screen key targets. The differential expression of candidate genes between LSCC and adjacent normal samples in TCGA database was tested using the Mann–Whitney U method, with the format of transcripts per million reads, followed by log2 conversion. R (version 3.6.3) “ggplot2” package was used to draw boxplots. The Kaplan–Meier survival curve of HRAS and SOX2 expression in lung cancer samples was retrieved from the KMplot database, and the patients were divided into two groups according to the median of gene expression, with p < 0.01 as the threshold.
Subsequently, genes interacting with SOX2 were predicted USING GeneCards and BioGRID databases, and the key upstream PAK2 was identified through PPI network analysis. The co-expression analysis of PAK2 and SOX2 in TCGA database was conducted utilizing the starBase tool. Moreover, the binding sites between them were predicted by the JASPAR database (Organisms: homo sapiens).
Ethics approval
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University. Detailed study aims as well as planned procedures were explained to all patients, who were subsequently provided with signed informed consent documentation. Animal experiments were approved by the Animal Care and Use Committee of the First Affiliated Hospital of Nanchang University and performed in accordance with Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Tissue collection and cell culture
LSCC tissues and adjacent normal tissues were collected from a total of 83 patients with LSCC who underwent initial surgery at the First Affiliated Hospital of Nanchang University from January 2015 to March 2017. These patients had never undergone preoperative chemotherapy or radiotherapy and received standard treatment after surgery in accordance with the 2004 National Comprehensive Cancer Network (NCCN) clinical practice guidelines in oncology: Non-Small Cell Lung Cancer. If the cancer recurred, subsequent treatment method was determined by the doctor in charged. All cases were classified following the World Health Organization criteria for LSCC.
Four LSCC cell lines (LTEP-S, SK-MES-1, NCI-H2170, NCI-H520), a normal lung epithelial cell line BEAS-2B and the normal human embryonic kidney HEK-293T cell line were purchased from the National Infrastructure of Cell Line Resource. BEAS-2B and LTEP-S cells were cultured in high-glucose DMEM (GIBCO, Grand Island, NY) containing 10% fetal bovine serum (FBS, GIBCO). NCI-H2170 and NCI-H520 cells were cultured in RPMI 1640 medium (GIBCO) containing 10% FBS. HEK-293T and SK-MES-1 cells were cultured in 10% FBS-containing MEM medium (GIBCO). All cells were cultured in a 5% CO2 incubator at 37 °C and 95% saturated humidity.
Immunohistochemistry (IHC)
The sections were immersed successively in xylene-absolute ethanol-95% ethanol-70% ethanol (5 min per time), followed by heating at 95 °C with 0.01 M sodium citrate buffer. The sections were then blocked with normal goat serum blocking solution at room temperature. Subsequently, the sections were incubated with corresponding primary antibodies (anti-SOX2, ab97959, 1:100; anti-PAK2, ab76293, 1:200; anti-DEK, ab249362, 1:100; Abcam, Cambridge, UK). Then, the sections were incubated with secondary antibody for 1 h at room temperature. After color development in DAB reagent, sections were treated with hematoxylin counterstaining for microscopic observation.
Cell transfection
LSCC cell lines NCI-H2170 and NCI-H520 were classified into 15 groups and subjected to different treatment with: short hairpin RNA (shRNA, sh)-negative control (NC), sh-SOX2-1, sh-SOX2-2, sh-SOX2-3, overexpression (oe)-NC, oe-SOX2, oe-PAK2 + sh-NC, oe-PAK2 + sh-SOX2, sh-PAK2, sh-DEK-1, sh-DEK-2, sh-DEK-3, sh-NC + oe-NC, sh-SOX2 + oe-NC, and sh-SOX2 + oe-DEK. The overexpression vector was pcDNA3.1, and the RNAi vector is pRNAT-U6.1/neo, both designed and constructed by GenePharma (Shanghai, China). The transfection was performed following the protocols of Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Cells were then incubated with G418 (400 ug/ml) to screen stable cells silencing DEK. Two weeks later, cells were incubated with G418 (200 ug/ml) again to screen stably transfected NCI-H2170 and NCI-H520 cells for two-month culture.
RNA extraction and quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was extracted by TRIzol reagent (15596026, Invitrogen) from tissues. Following the protocols of PrimeScript RT reagent Kit (RR047A, Takara, Shiga, Japan), the RNA was reverse-transcribed into cDNA. Then, SYBR Premix EX Taq kit (RR420A, Takara) and the ABI7500 PCR system (ABI, Foster City, CA) were used for qRT-PCR determination. Involved primers were synthesized by Shanghai Sangon Biotech (Shanghai, China), as listed in Supplementary Table 1. Each group was repeated in three wells. Further, the relative quantification method (2−△△CT method) was used to calculate the relative transcription level (normalized to β-actin).
Western blot assay
Total protein was extracted from cells utilizing RIPA lysis buffer containing phenylmethylsulphonyl fluoride, followed by determination of protein concentration using BCA detection kit (70-PQ0012, Multi Sciences, Hangzhou, Zhejiang, China). Then, the protein (50 µg) was separated by SDS-PAGE, electro-transferred to PVDF membrane, and blocked with 5% skim milk powder at room temperature for 1 h. Afterwards, the membrane was incubated overnight at 4 °C with diluted rabbit primary antibodies, including anti-DEK (ab74975, 1:500, Abcam), anti-Ki67 (ab16667, 1:1000, Abcam), anti-SOX2 (ab97959, 1:1000, Abcam), anti-MMP-9 (ab137867, 1:1000, Abcam), anti-E-cadherin (ab15148, 1:500, Abcam), anti-N-cadherin (ab18203, 1:1000, Abcam), and anti-β-actin (ab227387, 1:5000, Abcam). After washing, the membrane was further incubated for 1 h with horseradish peroxidase (HRP)-labeled IgG secondary antibody (ab97051, 1:2000, Abcam). The enhanced chemiluminescence detection kit (Thermo Fisher Scientific, San Jose, PA) was then utilized to visualize the protein bands. Further, the ECL fluorescence detection kit (BB-3501, Amersham, Little Chalfont, UK) was utilized to visualize the blot, and images were photographed with Bio-Rad image analysis system (BIO-RAD, Hercules, CA). The gray level of protein bands was quantified using Quantity One v4.6.2 software and normalized to β-actin.
Coimmunoprecipitation (Co-IP) assay
Cells were lysed with RIPA buffer (Thermo Fisher Scientific), sonicated for 30 min, and centrifuged at 13000 g for 30 min at 4 °C. The specific antibody and the supernatant were mixed overnight at 4 °C, followed by 4 h incubation with Pierce protein A/G Magnetic Beads (Thermo, 88803). Then, the beads were collected by centrifugation, mixed with loading buffer, and examined with SDS-PAGE and Western blot.
Glutathione-S-transferase (GST) pull-down assay
The CDS sequence of PAK2 was obtained from the NCBI Reference Sequence: NM_002577.4, constructed into Pgex-4T-3 plasmids by Beijing ABXBio to form a GST-PAK2 recombinant expression vector, and transformed into E. coli Rosetta. After positive cloning, prokaryotic expression was induced by IPTG, and the GST-PAK2 protein was purified by glutathione agarose affinity chromatography column. The purified GST-PAK2 protein and GST-labeled protein were then mixed with GST-Beads (Immobilized Glutathione) in an EP tube and rotated in a chromatography cabinet at 4 °C for 1 h. The cells transfected with plasmids overexpressing SOX2 were collected and fully lysed, and 20% of the lysate was taken as the input (positive control). The remaining lysate was then added to the EP tube and rotated in the chromatography cabinet at 4 °C for 4 h. Then, GST-Beads were washed and analyzed with SOX2 antibody in Western blot. Further, before washing, 2% volume of the mixture was taken for Western blot analysis.
Chromatin immunoprecipitation (ChIP) assay
The NCI-H2170 and NCI-H520 cells of each group were fixed with formaldehyde for 10 min to produce DNA-protein crosslinks and lysed with ultrasonic disruptor (each sonicate for 10 s, interval of 10 s, and 15 cycles) to break the chromatin into fragments (200–500 bp). Then, 10% of the cell lysate was set as the Input. The left lysate was centrifuged at 4 °C, 12000 g for 10 min to collect the supernatant, which were then divided into two tubes and respectively incubated overnight at 4 °C with the NC rabbits IgG (ab109489, 1:300, Abcam) and anti-SOX2 antibodies (Upstate Biotech, Lake Placid, NY). Afterwards, the DNA-protein complexes were precipitated with Pierce protein A/G Magnetic Beads (88803, Thermo Fisher Scientific), de-crosslinked, extracted with phenol/chloroform. Purified DNA fragments were measured with qRT-PCR assay.
After PAK2 knockdown, the sample containing purified DNA fragments was collected for ChIP assay (the same procedures as above) with sample of the sh-NC group as the control. Primers used to detect the enrichment of DEK promoter fragments was: Forward 5′-GATCTTTTTCCTTTCGGTG-3′ and Reverse 5′-TGCGTGTTTATTGTTTCCA-3′.
Immunofluorescence microscopy
Transfected NCI-H2170 and NCI-H520 cells were fixed with 4% polymethylene formaldehyde, 15 min permeabilization with 0.3% Triton-X100 and blocking with 5% BSA, cells were then incubated overnight with anti-SOX2 antibodies (1: 400; Cell Signaling Tech., Danvers, MA) at 4 °C, followed by incubation with the anti-rabbit immunoglobulin G secondary antibody that had been labeled by Alexa Fluor 488 (Beyotime, Beijing, China). Afterwards, cells were stained with DAPI (Sigma, St. Louis, MO) and then subjected to immunofluorescence microscopic observation using a CFD laser scanning microscope (DFC420C, LEICA, Wetzlar, Germany).
Phosphorylation assay
Cells were treated with lysis supplemented with protease/phosphatase inhibitors (Sigma), and dephosphorylated samples were then incubated for 1 h with the alkaline phosphatase (FastAP, Thermo Fisher Scientific), followed by separation of the protein utilizing Mn2+-Phos-TagTM SDS-PAGE as previously described15.
Cell counting kit-8 (CCK-8) assay
Cell proliferation was measured by Cell Counting Kit-8 kit (Dojindo, Kumamoto, Japan). Briefly, NCI-H2170 and NCI-H520 cells were seeded into a 96-well plate. Then, 10 μl of CCK-8 reagent was added into each well 24, 48, 72, and 96 h after transfection for 1 h incubation. Then, the absorbance at 450 nm was evaluated utilizing a microplate reader.
Transwell assays
Migration assay: LSCC cell suspension was added to the upper chamber of Transwell system and pre-chilled DMEM containing 10% FBS to the lower chamber. The chambers were incubated at 5% CO2 and 37 °C for 24 h. Then, cells in the lower chamber were fixed with methanol for 30 min, stained with 0.1% crystal violet reagent for 20 min, inverted, and dried. Five fields of view were randomly selected under an inverted microscope to count the number of cells penetrating the membrane.
Invasion assay: Diluted matrigel was added to the polycarbonate membrane of upper chamber. The following procedures were the same as the migration assay.
In vivo experiments
A total of 60 twenty-four-week-old BALB/C nude mice were purchased from the animal center of the First Affiliated Hospital of Nanchang University and randomly divided into six groups (ten mice/group). NCI-H2170 or NCI-H520 cells treated with sh-DEK-1, sh-DEK-2, or sh-NC were subcutaneously injected into the thighs of mice of the corresponding group. Then, the size of xenografted tumors was examined every week, and the volume was calculated following the formula: (a*b2)/2 (a: the longest diameter of the tumor; b: the shortest diameter of the tumor) and recorded. Six weeks after the injection, the nude mice were euthanized. The tumors were removed and weighed.
Statistical analysis
Data in this study was processed utilizing SPSS v.22.0 (SPSS Inc., Chicago, IL) software. Measurement data were summarized as mean ± standard deviation. Paired t-test was applied for the comparison of LSCC tissue and adjacent normal tissue, and unpaired t-test was applied for comparison between data of two groups. One-way analysis of variance (ANOVA) with Tukey’s post-hoc test was performed for comparison among data of multiple groups. The comparison of tumor volume between groups was performed by two-way ANOVA or repeated measurement ANOVA with Tukey’s post-hoc test. Pearson correlation analysis was adopted for the expression correlation of various factors in LSCC tissues. Kaplan–Meier was used to calculate the overall survival (OS) curve of patients with LSCC and log-rank to analyze the survival differences among patients. Cox regression analysis was applied for the prognosis-related factors in LSCC. Moreover, p < 0.05 indicated statistically significant difference.
Results
SOX2 is highly expressed in LSCC tissues and associated with poor prognosis of LSCC
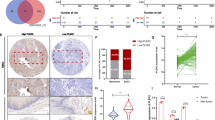
LSCC-related genes were retrieved from GeneCards (the top 300), DisGeNET (those with a score ≥ 0.1), and Phenolyzer databases, and the results of were then intersected to obtain 19 candidate genes (Fig. 1A). The expression of the 19 genes in LSCC (n = 502) and normal samples (n = 49) in TCGA database was then analyzed, and HRAS and SOX2 were highly expressed in LSCC tumor samples and the p value was the lowest (Fig. 1B). Results of the PPI network analysis further displayed that HRAS and SOX2 were at the core position of the network (Fig. 1C). The KMplot data indicated that only SOX2 was associated with poor prognosis of lung cancer patients, and lower survival rate was seen in lung cancer patients with high SOX2 expression (Fig. 1D). SOX2 was selected as the candidate gene.
A LSCC-related genes retrieved from GeneCards (https://www.genecards.org/), DisGeNET (http://www.disgenet.org/search), and Phenolyzer (http://phenolyzer.wglab.org/) databases; B A box plot of the expression of 19 candidate genes in LSCC samples (n = 502) and normal samples (n = 49) in TCGA database. C PPI network of LSCC-related genes; the color of the node changing from blue to yellow indicates the Degree value of genes from high to low. D Kaplan–Meier curve analysis of the correlation of SOX2 with the prognosis of lung cancer patients in the KMplot database; red line indicates SOX2 high expression and black line indicates SOX2 low expression. The patients were grouped based on median expression. E qRT-PCR to measure the expression of SOX2 in LSCC tissues and adjacent normal tissues (n = 83), *p < 0.05 versus adjacent normal tissues; F qRT-PCR to determine SOX2 expression in LSCC tissues of different TNM stage (29 in Stage I, 32 in Stage II, and 22 in Stage III), *p < 0.05 versus Stage I tissues, #p < 0.05 versus Stage II tissues; G Kaplan–Meier curve analysis of the relationship between SOX2 expression and the OS and DFS of LSCC patients, and Log-rank method for differential analysis of survival rate; H IHC assay to determine the expression of SOX2 in representative LSCC tissues and adjacent normal tissues; I qRT-PCR and Western blot assay to measure the expression of SOX2 in four LSCC cell lines and a normal lung epithelial cell line. *p < 0.05 versus BEAS-2B cells. Measurement data were summarized as mean ± standard deviation. Paired t-test was applied for comparison between LSCC tissues and adjacent normal tissues (for D, E, H). One-way ANOVA with Tukey’s post hoc test was adopted for comparison of data of multiple groups (for F, I). Cell experiment was repeated three times.
In order to verify these results, we examined SOX2 expression in LSCC tissues and adjacent normal tissues of 83 LSCC patients by qRT-PCR. The expression of SOX2 was elevated in LSCC tissues compared to adjacent tissues (Fig. 1E), and LSCC of higher stage was associated with higher expression of SOX2 (Fig. 1F). We further evaluated the relationship between SOX2 expression and clinicopathological factors of the 83 patients (Supplementary Table 2), and a positive correlation between SOX2 expression and TNM stage of LSCC (also lymph node metastasis) was revealed. SOX2 expression was not correlated with other clinicopathological factors. Moreover, Kaplan–Meier curve analysis indicated that LSCC patients with higher SOX2 expression tended to have shorter OS and disease-free survival (DFS) (Fig. 1G).
IHC results also validated the up-regulated expression of SOX2 in representative LSCC tissue relative to adjacent normal tissues (Fig. 1H and Supplementary Fig. 1A). Moreover, through univariate analysis we further revealed that SOX2 expression, T staging, TNM stage, and lymph node metastasis were significantly related to OS, while multivariate analysis identified them as independent prognostic factors for LSCC patients (Supplementary Table 3). Further, we determined SOX2 expression in four LSCC cell lines (LTEP-S, SK-MES-1, NCI-H2170, and NCI-H520) and a normal lung epithelial cell line (BEAS-2B) by qRT-PCR and Western blot assays (Fig. 1I and Supplementary Fig. 2A). The results showed that SOX2 expression in LSCC cells was augmented versus that in BEAS-2B cells, and the highest expression of SOX2 was identified in NCI-H2170 and NCI-H520 cells, which were thus selected for subsequent experiments.
SOX2 knockdown represses the proliferative, migratory and invasive potential of LSCC cells
After identifying the up-regulated expression of SOX2 in LSCC tissues and cells, we further explored the effect of SOX2 dysregulation on LSCC cells. Constructing three shRNAs targeting SOX2 and transfecting them into NCI-H2170 and NCI-H520 cells, we then identified sh-SOX2-1 and sh-SOX2-2 as the two sh-RNAs with the best or second best SOX2-silencing efficiency utilizing qRT-PCR and Western blot (Fig. 2A and Supplementary Fig. 2B). Herein, sh-SOX2-1 and sh-SOX2-2 were selected for subsequent transfection of LSCC cells (NCI-H2170 and NCI-H520).
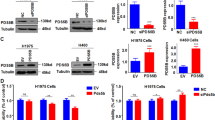
A qRT-PCR and Western blot assay to measure the silencing efficiency of three shRNAs targeting SOX2; B CCK-8 assay to evaluate the viability of NCI-H2170 and NCI-H520 cells transfected with sh-SOX2-1 and sh-SOX2-2; C Transwell assay to examine the migratory and invasive ability of NCI-H2170 and NCI-H520 cells transfected with sh-SOX2-1 and sh-SOX2-2; D Western blot determination of proliferation and migration-related proteins (Ki67, MMP-9, E-cadherin, and N-cadherin) in NCI-H2170 and NCI-H520 cells transfected with sh-SOX2-1 and sh-SOX2-2. Measurement data were summarized as mean ± standard deviation. One-way ANOVA with Tukey’s post hoc test was adopted for comparison among data of multiple groups (for A, C, D). Two-way ANOVA with Tukey’s post hoc test was used for comparison between data of two groups at different time points (for B). * p < 0.05 versus the sh-NC group. Cell experiment was repeated three times.
Following successful transfection, we adopted CCK-8 assay to measure the viability of transfected NCI-H2170 and NCI-H520 cells (Fig. 2B). According to the results, SOX2 knockdown, induced by either sh-SOX2-1 or sh-SOX2-2, obviously repress the viability of LSCC cells. Then, through Transwell assay we substantiated that SOX2 knockdown could suppress the migratory and invasive ability of LSCC cells (Fig. 2C and Supplementary Fig. 3A). Further, we determined the levels of a proliferation-related protein Ki67 and migration-related proteins (MMP-9, E-cadherin, and N-cadherin) by Western blot (Fig. 2D, Supplementary Fig. 2C), and the results showed that silencing SOX2 attenuated cell invasion and migration in LSCC cells. Taken together, SOX2 knockdown was able to repress the viability and the migratory and invasive potential of LSCC cells.
SOX2 directly interacts with PAK2
The aforementioned experiments have substantiated the regulatory effect of SOX2 on the biological activities of LSCC cells, whereas the underlying mechanisms remained unclear. In this sense, we searched SOX2-interacting proteins through GeneCards and BioGRID databases (screening score ≥ 0.9) and obtained 12 proteins (Fig. 3A). PPI network analysis was then performed on these proteins, among which PAK2, PAK1, OXSM, DDAH2, and PRTEDC1 were highlighted for their close interaction (Fig. 3B). Moreover, GEPI2 analysis indicated that PAK2 expression was up-regulated in LSCC tissues from TCGA database (Fig. 3C). Further, starBase analysis revealed that PAK2 and SOX2 expression was positively correlated in LSCC tissues from TCGA database (Fig. 3D).
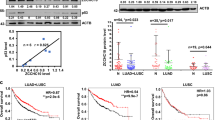
A SOX2-interacting proteins obtained from GeneCards and BioGRID (https://thebiogrid.org/) databases; B PPI network of SOX2-interacting proteins; C GEPIA2 analysis (http://gepia2.cancer-pku.cn/#index) on the differential expression of PAK2 in LSCC tissues and adjacent normal tissues; D starBase analysis (http://starbase.sysu.edu.cn/) of the correlation between the expression of PAK2 and SOX2 in LSCC tissues from the TCGA database; E Pearson correlation analysis of PAK2 and SOX2 expression in 83 cases of LSCC tissues; F IHC assay to determine the expression of PAK2 in LSCC tissues and adjacent normal tissues (*p < 0.05 versus adjacent normal tissues); G Co-IP assay to examine the binding of exogenous PAK2 and SOX2 proteins in HEK293T cells; H Co-IP assay to examine the binding of endogenous PAK2 and SOX2 proteins in NCI-H2170 and NCI-H520 cells; I GST pull-down assay in NCI-H2170 and NCI-H520 cells to validate the direct interaction between PAK2 and SOX2. J Mn2+-Phos-TagTM SDS-PAGE analysis of the phosphorylation of SOX2 protein (AP, alkaline phosphatase); K Immunofluorescence staining to detect the expression of SOX2 in cell nucleus (DAPI in blue. SOX2 in green). *p < 0.05 versus the sh-NC or the oe-NC group. Measurement data were summarized as mean ± standard deviation. Paired t-test was used for comparison between LSCC tissues and adjacent normal tissues (for C, F). Pearson correlation analysis was adopted for the expression correlation of various factors in LSCC tissues. Cell experiment was repeated three times.
To further verify their interaction, we adopted Pearson correlation analysis on the expression of PAK2 and SOX2 in 84 LSCC cases and identified the positive correlation between the expression of PAK2 and SOX2 in LSCC (Fig. 3E). In addition, IHC measurement revealed that PAK2 was highly expressed in LSCC tissues (Fig. 3F and Supplementary Fig. 1B). Moreover, we transferred exogenous PAK2 and SOX2 into HEK293T cells and performed Co-IP assay. The results indicated that PAK2 interacted with SOX2 (Fig. 3G). Next, we performed Co-IP assay of PAK2 and SOX2 in NCI-H2170 and NCI-H520 cells and found that endogenous PAK2 also interacts with SOX2 (Fig. 3H). Subsequently, GST pull-down assay (Fig. 3I) substantiated the direct interaction between PAK2 and SOX2. Moreover, silencing PAK2 led to reduced levels of phosphorylated SOX2 in NCI-H2170 and NCI-H520 cells (Fig. 3J). Consistently, immunofluorescence results showed that SOX2 expression was attenuated in response to PAK2 knockdown and was up-regulated in response to PAK2 overexpression (Fig. 3K). Taken together, these results indicated that SOX2 interacted with PAK2.
SOX2 participates in the stimulating effect of PAK2 on malignant phenotypes of LSCC cells
Following identification of the direct interaction between PAK2 and SOX2, we then explored the role of such interaction in the biological characteristics of LSCC cells. With this purpose, we established NCI-H2170 and NCI-H520 cell models of PAK2 overexpression as well as PAK2 overexpression + SOX2 knockdown, and then validated the models through qRT-PCR and Western blot assays (Fig. 4A and Supplementary Fig. 2D). Next, results of CCK-8 and Transwell assays indicated augmented proliferation, migration and invasion of LSCC cells in the presence of PAK2 overexpression alone (shown in the oe-PAK2 + sh-NC group versus the oe-NC + sh-NC group), whereas its combination with SOX2 knockdown resulted in the opposite results, as reflected by the reduced functions in the oe-PAK2 + sh-SOX2 group relative to the oe-PAK2 + sh-NC group (Fig. 4B, C and Supplementary Fig. 3B). Further, the expression levels of a proliferation-related protein Ki67 and migration-related proteins (MMP-9, E-cadherin, and N-cadherin) in LSCC cells were determined by Western blot. According to the results, PAK2 overexpression resulted in up-regulated levels of Ki67, MMP-9, and N-cadherin as well as down-regulated level of E-cadherin, whereas the combination of PAK2 overexpression and SOX2 knockdown resulted in the opposite (Fig. 4D and Supplementary Fig. 2E). In summary, PAK2 could stimulate the proliferative, migratory, and invasive abilities of LSCC cells only in the presence of SOX.
A qRT-PCR and Western blot assay to measure the expression of PAK2 and SOX2 in response to oe-PAK2 alone or combined with sh-SOX2; B CCK-8 assay to measure the proliferative ability of NCI-H2170 and NCI-H520 cells in response to oe-PAK2 alone or combined with sh-SOX2; C Transwell assay to detect the migratory and invasive potential of NCI-H2170 and NCI-H520 cells in response to oe-PAK2 alone or combined with sh-SOX2; D Western blot to determine the expression of Ki67, MMP-9, E-cadherin, and N-cadherin in NCI-H2170 and NCI-H520 cells in response to oe-PAK2 alone or combined with sh-SOX2. Measurement data were summarized as mean ± standard deviation. One-way with Tukey’s post hoc test was adopted for comparison among data of multiple groups (for A, C, D). Two-way ANOVA with Tukey’s post hoc test was used for comparison among data at different time points (for B). *p < 0.05 versus the oe-NC + sh-NC group; #p < 0.05 versus the oe-PAK2 + sh-NC. Cell experiment was repeated three times.
PAK2 and SOX2 co-regulates the downstream DEK gene
Recent studies show that SOX2 had a role to confer in SCC via up-regulating the expression of DEK, ETV4, HRAS, SRSF2, JUN, YAP1, SOX2OT, CDH1 and other genes5,16,17,18. Herein, we conducted a quantitative analysis on these genes. The results indicated that DEK presented the most obvious up-regulation of expression in the LSCC cells (LTEP-S, SK-MES-1, NCI-H2170, NCI-H520) versus the normal lung epithelial cell line (BEAS-2B) (Fig. 5A). IHC results also showed the elevated expression of DEK in LSCC tissues (Fig. 5B, Supplementary Fig. 1C). Then the JASPAR database was utilized to predict the binding sites of SOX2 and DEK (Fig. 5C).
A qRT-PCR to determine the expression of DEK in human LSCC cells and normal lung epithelial cells, *p < 0.05 versus BEAS-2B cells; B IHC assay to determine the expression of DEK in LSCC tissues and adjacent normal tissues; *p < 0.05 versus adjacent normal tissues; C Binding sites of SOX2 and DEK predicted by the JASPAR database (http://jaspar.genereg.net/); D ChIP assay to examine the binding of SOX2 to DEK and SOX1 promoter region, *p < 0.05 versus IgG antibody group; E qRT-PCR to determine the expression of DEK in cells, *p < 0.05 versus the oe-NC or oe-NC + sh-NC group; F qRT-PCR and Western blot to measure the silencing efficiency of sh-PAK2 and DEK expression upon sh-PAK2 treatment; *p < 0.05 versus the sh-NC group; G ChIP assay to examine the binding of SOX2 and DEK promoter region after PAK2 knockdown, *p < 0.05 versus the sh-NC or oe-NC group; H Co-expression analysis of SOX2 and DEK in LSCC samples in TCGA database; I Co-expression analysis of PAK2 and DEK in LSCC samples in TCGA database. Measurement data were summarized as mean ± standard deviation. One-way ANOVA with Tukey’s post hoc test was adopted for comparison among data of multiple groups (for A, E). Paired t-test was used for comparison between LSCC tissues and adjacent normal tissues (for B), while unpaired t-test was used for comparison between data of two groups (for D, F, G). Cell experiment was repeated three times.
Furthermore, we conducted ChIP assays in NCI-H2170 and NCI-H520 cells and identified that SOX2 could bind to the promoter region of DEK (Fig. 5D). According to the published SOX2 genomic binding map19,20, we selected the SOX1 promoter as a NC and examined the binding of SOX2 to SOX1 promoter by ChIP assay in NCI-H2170 and NCI-H520 cells, respectively. The results displayed that the amount of SOX1 promoter fragments enriched by the SOX2 antibody group has no obvious difference with that of SOX1 promoter fragments enriched by the IgG antibody (Fig. 5D), indicating that SOX2 cannot bind to SOX1 promoter region. Next, qRT-PCR results showed that SOX2 overexpression could lead to elevated expression of DEK in both NCI-H2170 and NCI-H520 cells (Fig. 5E). Subsequently, through qRT-PCR we measured DEK expression in NCI-H2170 and NCI-H520 cell model of PAK2 overexpression and PAK2 overexpression + SOX2 knockdown (Fig. 5E). According to the results, the expression of DEK was elevated in response to PAK2 overexpression alone, but decreased in response to the combination of PAK2 overexpression and SOX2 knockdown.
Further, we treated NCI-H2170 and NCI-H520 cells with sh-PAK2, and the expression of PAK2 in these cells was successfully knocked down, as shown by qRT-PCR and Western blot measurement; PAK2 knockdown was then observed to result in reduced expression of DEK (Fig. 5F and Supplementary Fig. 2F). We then investigated the binding of SOX2 and DEK promoter region by ChIP assays. As a result, the DEK promoter region fragments enriched by SOX2 antibodies were obviously reduced in response to PAK2 knockdown and increased in response to PAK2 overexpression (Fig. 5G), indicating that PAK2 knockdown repressed SOX2 binding to the promoter region of DEK. Then, through GEPIA2 analysis we further validated that DEK expression was positively correlated with SOX2 and PAK2 expression in LSCC tissues from the TCGA database (Fig. 5H, I). Taken together, PAK2 and SOX2 could co-regulate DEK.
DEK knockdown represses the malignant phenotypes of LSCC cells
The previous experiments revealed the effect of SOX2 and PAK2 on the biological activities of LSCC cells and identified DEK as the target gene of SOX2 and PAK2. We speculated that DEK might also have regulatory effect on LSCC cells. To validate this hypothesis, we designed and constructed three shRNAs targeting DEK and transfected them into NCI-H2170 and NCI-H520 cells. Then, through qRT-PCR and Western blot, we identified the sh-DEK-1 and sh-DEK-2 with the best and second-best silencing efficiency (Fig. 6A and Supplementary Fig. 2G) and selected them for transfection of LSCC cells (NCI-H2170 and NCI-H520 cells).
A qRT-PCR and Western blot to determine the silencing efficiency of three shRNAs targeting DEK; B CCK-8 assay to measure the proliferative ability of NCI-H2170 and NCI-H520 cells transfected with sh-DEK-1 and sh-DEK-2; C Transwell assay to determine the invasive and migratory abilities of NCI-H2170 and NCI-H520 cells transfected with sh-DEK-1 and sh-DEK-2; D Western blot to determine the levels of Ki67, MMP-9, E-cadherin, and N-cadherin in NCI-H2170 and NCI-H520 cells transfected with sh-DEK-1 and sh-DEK-2; E qRT-PCR and Western blot to measure the expression of DEK in the tissues of xenografted tumors; F The growth curve to reflect the volume of xenografted tumors in mice; G Weight of xenografted tumors; H Western blot to measure the levels of Ki67, MMP-9, E-cadherin, and N-cadherin in the tissues of xenografted tumors. Ten mice in each group. Measurement data were summarized as mean ± standard deviation. One-way ANOVA with Tukey’s post hoc test was adopted for comparison among data of multiple groups (for A, C, D, E, G, H). Two-way (for B) or repeated measurement (for F) ANOVA with Tukey’s post hoc for comparison among multiple groups at different time points. *p < 0.05 versus the sh-NC group. Cell experiment was repeated three times.
The viability and the migratory and invasive potential of cells transfected with sh-DEK-1 or sh-DEK-2 were evaluated with CCK-8 and Transwell assays (Fig. 6B, C), which revealed that DEK knockdown could obviously repress malignant phenotypes of LSCC cells. Moreover, we adopted Western blot to measure expression levels of a proliferation-related protein Ki67 and migration-related proteins (MMP-9, E-cadherin, and N-cadherin) in NCI-H2170 and NCI-H520 cells, and the results revealed that silencing DEK repressed the expression of Ki67, MMP-9 and N-cadherin and promote the expression of anti-migratory E-cadherin (Fig. 6D and Supplementary Fig. 2H). In addition, our data demonstrate that SOX2 knockdown reduced cell viability, attenuated cell migration and invasion, down-regulated the expression of Ki67, MMP-9 and N-cadherin, and up-regulated E-cadherin expression in NCI-H2170 and NCI-H520 cells; while additional DEK overexpression could reverse the aforementioned effects of SOX2 knockdown alone (Supplementary Fig. 4A, B, C).
In order to further explore the effect of DEK on the tumorigenesis of LSCC in vivo, we established NCI-H2170 and NCI-H520 cells silencing DEK and injected them into nude mice. Then, we determined the expression of DEK by qRT-PCR and Western blot, the results of which identified that the expression of DEK in xenografted tumors was decreased in response to sh-DEK treatment (Fig. 6E). Further, as shown in Supplementary Fig. 5 and Fig. 6F, G, DEK knockdown resulted in reduced volume and weight of xenografted tumors. Moreover, Western blot analysis in the xenografted tumor tissues indicated that the levels of Ki67, MMP-9 and N-cadherin were down-regulated and the level of E-cadherin was up-regulated in the presence of DEK knockdown (Fig. 6H and Supplementary Fig. 2I). In summary, DEK knockdown could repress the viability and the migratory, and invasive potential of LSCC cells both in vitro and in vivo.
Discussion
In this study, we show that PAK2 contributes to the development of LSCC by positively interacting with SOX2 and up-regulating the expression of oncogenic DEK (Fig. 7).
SOX2 is a transcription factor modulating pluripotent stem cells, and the amplification of SOX2 has been reported in a spectrum of SCCs including esophageal SCC21 and head and neck cutaneous SCC22. More importantly, accumulating data have revealed that dysregulated SOX2 was involved in the formation of LSCC23,24. In agreement with these reports, our bioinformatic analysis identified the overexpression of SOX2 in LSCC tissues and correlated it with the poor prognosis of LSCC. Further, through a series of functional assays we substantiated that SOX2 knockdown was able to repress the viability and the migratory and invasive potential of LSCC cells. Such regulatory effects on cell functions, as a previous study has delineated, may attribute to the role of SOX2 as a driver gene for the recurrent 3q26.33 (one of the most frequently amplified genomic sites in SCC) amplifications in LSCC25. In this study, we further explored the downstream molecular mechanism and identified PAK2 as a SOX2-interacting gene. This finding corroborates a previous research where PAK2 was indicated to interact with SOX29. Furthermore, our data revealed that PAK2 directly bound to SOX2. Herein, it was reasonable to hypothesize that PAK2 had a role to confer in the tumorigenesis of LSCC.
The transcription factor SOX2 is involved in a variety of biological processes, including maintenance of embryonic and adult stem cells, bronchial and lung differentiation, tumorigenesis and cell survival26,27,28,29,30. Recent data have documented that SOX2 has an important role to play in LUSC tumor stem cell phenotypes15. Indeed, amplification of SOX2, consistent with its overexpression, is a common genomic alteration in LUSC of TCGA cohort31. Corroborating findings are identified in a prior study, which demonstrated that ~75% of LUSC patients in China were found to carry SOX2 amplification32. Our work also found that SOX2 overexpression occurred in LSCC. However, the association between SOX2 and indicators of better prognosis in LSCC should also be noted in a previous study, where increased SOX2 may prolong the survival of patients with LSCC8. Additionally, a study by Lu et al. found that the prognosis of patients with high SOX2 expression may be more satisfactory than those with low SOX2 expression33, but this point could not be replicated using the Michigan squamous cell carcinoma samples. In addition, some researchers found that the prognosis of patients with high SOX2 expression was better than those with low SOX2 expression, but they also pointed out that the SOX2 expression was appreciably correlated with the pathological type of cancer34,35,36. Previous studies suggest that LSCC patients with high SOX2 expression might present better prognosis. Nevertheless, our results indicated that SOX2 may be carcinogenic during lung cancer development, which may be due to the difference in methods employed by different laboratories, such as molecular methods for detecting SOX2 copy number status, or threshold settings for distinguishing different amplification levels. An additional consideration is the difference between cohort and tumor heterogeneity.
PAKs belong to the non-receptor serine/threonine protein kinase family and have been well studied for locating at the intersection of several signaling pathways that mediate multiple cellular processes and participating in cancer development37. In this study we identified the up-regulated expression of PAK2 in LSCC tissues and substantiated that PAK2 could trigger malignant behaviors of LSCC cells. Furthermore, through a series of gain- and loss- of function assays we uncovered that the existence of SOX2 was the precondition for the regulatory effect of PAK2 on LSCC cells. In other words, PAK2 modulated the formation of LSCC through interacting SOX2. Subsequently, we managed to explore the downstream mechanism and found that PAK2 and SOX2 co-regulated DEK gene. As a typical oncogene, DEK has been reported for being up-regulated in various cancers11. Furthermore, emerging evidence has revealed that the overexpression of DEK in NSCLC participated in the pathogenesis of NSCLC via stimulating the proliferation and invasion of NSCLC cells12,13,14. In agreement with these reports, our data substantiated that the knockdown of DEK could repress the malignant phenotypes of LSCC cells. Our in vivo experiment further verified the LSCC-inhibiting function of DEK knockdown. Considering DEK was co-modulated by SOX2 and PAK2, in this study we identified the PAK2/SOX2/DEK axis involved in the pathogenesis of LSCC.
In conclusion, PAK2 binds to the transcription factor SOX2 and thus activates the expression of DEK, thereby driving malignant behavior and tumor growth of LSCC cells. The overexpression of PAK2 was able to augment the viability and the migratory and invasive potential of LSCC cells, while the knockdown of SOX2 or DEK reversed the promoting effect of PAK2 overexpression. Our study lays a theoretical foundation for deeper understanding of the pathogenesis of LSCC. More importantly, our findings provide new therapeutic targets and insightful information for enhancing targeted therapy of LSCC.
Data availability
The data that supports the findings of this study are available in the paper and supplementary materials.
References
Spira A, Halmos B, Powell CA. Update in Lung Cancer 2015. Am J Respir Crit Care Med. 2016;194:661–71.
Mansour H, Ouhajjou A, Bajic VB, Incitti R. Next-Generation Sequencing at High Sequencing Depth as a Tool to Study the Evolution of Metastasis Driven by Genetic Change Events of Lung Squamous Cell Carcinoma. Front Oncol. 2020;10:1215.
Liao RG, Watanabe H, Meyerson M, Hammerman PS. Targeted therapy for squamous cell lung cancer. Lung Cancer Manag. 2012;1:293–300.
Friedlaender A, Banna G, Malapelle U, Pisapia P, Addeo A. Next Generation Sequencing and Genetic Alterations in Squamous Cell Lung Carcinoma: Where Are We Today? Front Oncol. 2019;9:166.
Watanabe H, Ma Q, Peng S, Adelmant G, Swain D, Song W, et al. SOX2 and p63 colocalize at genetic loci in squamous cell carcinomas. J Clin Investig. 2014;124:1636–45.
Gen Y, Yasui K, Zen Y, Zen K, Dohi O, Endo M, et al. SOX2 identified as a target gene for the amplification at 3q26 that is frequently detected in esophageal squamous cell carcinoma. Cancer Genet Cytogenet. 2010;202:82–93.
Maier S, Wilbertz T, Braun M, Scheble V, Reischl M, Mikut R, et al. SOX2 amplification is a common event in squamous cell carcinomas of different organ sites. Hum Pathol. 2011;42:1078–88.
Wilbertz T, Wagner P, Petersen K, Stiedl AC, Scheble VJ, Maier S, et al. SOX2 gene amplification and protein overexpression are associated with better outcome in squamous cell lung cancer. Mod Pathol. 2011;24:944–53.
Huttlin EL, Ting L, Bruckner RJ, Gebreab F, Gygi MP, Szpyt J, et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015;162:425–40.
Wang Y, Tong J, Lin H, Ma L, Xu Y. CCHE1 accelerated the initiation of oral squamous cell carcinoma through enhancing PAK2 expression by sponging miR-922. J Oral Pathol Med. 2020;49:636–44.
Yang MQ, Bai LL, Lei L, Zheng YW, Wang Z, Li ZH, et al. DEK promotes the proliferation and invasion of lung cancers and indicates poor prognosis in lung adenocarcinomas. Oncol Rep. 2020;43:1338–48.
Liu X, Qi D, Qi J, Mao Z, Li X, Zhang J, et al. Significance of DEK overexpression for the prognostic evaluation of non-small cell lung carcinoma. Oncol Rep. 2016;35:155–62.
Qian Z, Yang J, Liu H, Yin Y, Hou L, Hu H. The miR-1204 regulates apoptosis in NSCLC cells by targeting DEK. Folia Histochem Cytobiol. 2019;57, 64–73.
Zhou QC, Deng XF, Yang J, Jiang H, Qiao MX, Liu HH, et al. Oncogene DEK is highly expressed in lung cancerous tissues and positively regulates cell proliferation as well as invasion. Oncol Lett. 2018;15:8573–81.
Bao Z, Ji W, Yang Y, Chen Z, Li Z, Wang K, et al. PAK5 promotes the cell stemness ability by phosphorylating SOX2 in lung squamous cell carcinomas. Exp Cell Res. 2020;395:112187.
Chai Y, Li Q, Zhao H, Zhang Z, Yu X, Pang L, et al. SOX2 antagonizes WWC1 to drive YAP1 activation in esophageal squamous cell carcinoma. Cancer Med. 2019;8:7055–64.
Wu Y, Chen X, Liang Y, Li J, Zhang K, Dai L, et al. Overexpression of long non-coding RNA SOX2OT promotes esophageal squamous cell carcinoma growth. Cancer Cell Int. 2018;18:76.
Keysar SB, Le PN, Miller B, Jackson BC, Eagles JR, Nieto C, et al. Regulation of Head and Neck Squamous Cancer Stem Cells by PI3K and SOX2. J Natl Cancer Inst. 2017;109:djw189. https://doi.org/10.1093/jnci/djw189.
Zhou C, Yang X, Sun Y, Yu H, Zhang Y, Jin Y. Comprehensive profiling reveals mechanisms of SOX2-mediated cell fate specification in human ESCs and NPCs. Cell Res. 2016;26:171–89.
Zimmerman DL, Boddy CS, Schoenherr CS. Oct4/Sox2 binding sites contribute to maintaining hypomethylation of the maternal igf2/h19 imprinting control region. PLoS ONE. 2013;8, e81962.
Tan Y, Lu X, Cheng Z, Pan G, Liu S, Apiziaji P, et al. miR-148a Regulates the Stem Cell-Like Side Populations Distribution by Affecting the Expression of ACVR1 in Esophageal Squamous Cell Carcinoma. Onco Targets Ther. 2020;13:8079–94.
Kilmister EJ, Patel J, van Schaijik B, Bockett N, Brasch HD, Paterson E, et al. Cancer Stem Cell Subpopulations Are Present Within Metastatic Head and Neck Cutaneous Squamous Cell Carcinoma. Front Oncol. 2020;10:1091.
Liu Y, Yin N, Wang X, Khoor A, Sambandam V, Ghosh AB, et al. Chromosome 3q26 Gain Is an Early Event Driving Coordinated Overexpression of the PRKCI, SOX2, and ECT2 Oncogenes in Lung Squamous Cell Carcinoma. Cell Rep. 2020;30:771–82 e6.
Wang Y, Tan X, Tang Y, Zhang C, Xu J, Zhou J, et al. Dysregulated Tgfbr2/ERK-Smad4/SOX2 Signaling Promotes Lung Squamous Cell Carcinoma Formation. Cancer Res. 2019;79:4466–79.
Hussenet T, Dali S, Exinger J, Monga B, Jost B, Dembele D, et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplifications in human lung squamous cell carcinomas. PLoS ONE 2010;5:e8960.
Novak D, Huser L, Elton JJ, Umansky V, Altevogt P, Utikal J. SOX2 in development and cancer biology. Semin Cancer Biol. 2020;67:74–82.
Boumahdi S, Driessens G, Lapouge G, Rorive S, Nassar D, Le Mercier M, et al. SOX2 controls tumour initiation and cancer stem-cell functions in squamous-cell carcinoma. Nature. 2014;511:246–250.
Feng R, Wen J. Overview of the roles of Sox2 in stem cell and development. Biol Chem. 2015;396:883–891.
Liu K, Lin B, Zhao M, Yang X, Chen M, Gao A, et al. The multiple roles for Sox2 in stem cell maintenance and tumorigenesis. Cell Signal. 2013;25:1264–1271.
Batchuluun K, Azuma M, Fujiwara K, Yashiro T, Kikuchi M. Notch Signaling and Maintenance of SOX2 Expression in Rat Anterior Pituitary Cells. Acta Histochem Cytochem. 2017;50:63–69.
Zengin T, Onal-Suzek T. Comprehensive Profiling of Genomic and Transcriptomic Differences between Risk Groups of Lung Adenocarcinoma and Lung Squamous Cell Carcinoma. J Pers Med. 2021;11:154. https://doi.org/10.3390/jpm11020154.
Tan Q, Li F, Wang G, Xia W, Li Z, Niu X, et al. Identification of FGF19 as a prognostic marker and potential driver gene of lung squamous cell carcinomas in Chinese smoking patients. Oncotarget. 2016;7:18394–18402.
Lu Y, Futtner C, Rock JR, Xu X, Whitworth W, Hogan BL, et al. Evidence that SOX2 overexpression is oncogenic in the lung. PLoS ONE 2010;5:e11022.
Chen Y, Huang Y, Huang Y, Chen J, Wang S, Zhou J. The prognostic value of SOX2 expression in non-small cell lung cancer: a meta-analysis. PLoS ONE 2013;8:e71140.
Ying J, Shi C, Li CS, Hu LP, Zhang WD. Expression and significance of SOX2 in non-small cell lung carcinoma. Oncol Lett. 2016;12:3195–3198.
Velcheti V, Schalper K, Yao X, Cheng H, Kocoglu M, Dhodapkar K, et al. High SOX2 levels predict better outcome in non-small cell lung carcinomas. PLoS ONE 2013;8:e61427.
Wang J, Zhu Y, Chen J, Yang Y, Zhu L, Zhao J, et al. Identification of a novel PAK1 inhibitor to treat pancreatic cancer. Acta Pharm Sin B. 2020;10:603–14.
Author information
Authors and Affiliations
Contributions
SX conceived and designed research. XW performed experiments and interpreted results of experiments. SC analyzed data and prepared figures. YH drafted paper. XL edited and revised paper. All authors read and approved final version of paper. This study was supported by National Natural Science Foundation of China (No. 82060507) and Natural Science Foundation of Jiangxi Province (No. 20202BABL206074).
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the First Affiliated Hospital of Nanchang University. Detailed study aims as well as planned procedures were explained to all patients, who were subsequently provided with signed informed consent documentation. Animal experiments were approved by the Animal Care and Use Committee of the First Affiliated Hospital of Nanchang University and performed in accordance with Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Xie, S., Wan, X., Chen, S. et al. p21-activated kinase 2 binds to transcription factor SOX2 and up-regulates DEK to promote the progression of lung squamous cell carcinoma. Lab Invest 102, 1109–1120 (2022). https://doi.org/10.1038/s41374-022-00808-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-022-00808-5