Abstract
Both toll-like receptors (TLRs) and nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs) induce a tightly regulated inflammatory response at risk of causing tissue damage, depending on the effectiveness of ensuing negative feedback regulatory mechanisms. Cross-regulation between TLRs, NLRs, and cytokine receptors has been observed. However, the cross-regulation between interleukin-1 (IL-1) receptors and NOD2 is not completely understood. In this study, we found that IL-1α/β increased NOD2-induced inflammatory response in human monocytic THP1 cells, peripheral blood mononuclear cells (PBMCs), mouse macrophage RWA264.7 cells and spleen cells, and in an in vivo experiment. IL-1α/β pre-treatment induced the production of CXC chemokines, including growth-regulated oncogene (GRO)-α, GRO-β, and IL-8, and proinflammatory cytokines, including IL-1β, IL-6, and TNFα, which are induced by the activation of NOD2, in a dose- and time-dependent manner. However, pre-treatment with the NOD2 ligand muramyl dipeptide (MDP) did not up-regulate the expression of cytokines induced by IL-1α/β re-treatment. IL-1β treatment increased the expression of A20, which is an important inhibitor of the innate immune response. However, the overexpression of A20 failed to inhibit MDP-induced cytokine production, suggesting that A20 had no effects on the NOD2-induced immune response. In addition, IL-1α/β increased the expression of NOD2 and its downstream adaptor RIP2, and IL-1α/β pre-treatment increased MDP-induced activation of mitogen-activated protein kinases (MAPKs), including ERK, JNK, and P38, which contributed to MDP-induced cytokine production. Based on these results, IL-1α/β promote the NOD2-induced immune responses by enhancing MDP-induced activation of MAPK signaling pathways.
Similar content being viewed by others
Introduction
The innate immune system constitutes the first line for the defense against invading pathogens [1]. Recognition of conserved microbial molecules named pathogen- associated molecular patterns (PAMPs) by pattern recognition receptors (PRRs), such as transmembrane toll-like receptors (TLRs) or cytosolic nucleotide-binding and oligomerization domain (NOD)-like receptors (NLRs), generally results in the production of inflammatory mediators, subsequent recruitment and activation of leukocytes, and the initiation of the inflammatory responses [1].
NOD2 is one of the earliest NLRs characterized. The NLR family comprises 22 proteins in humans. All members have a characteristic tri-domain structure consisting of a C-terminal leucine-rich repeat (LRR) domain, a central nucleotide-binding and oligomerization (NACHT) domain, and an N-terminal effector domain [2]. The LRR domain functions in ligand recognition. The central NACHT domain facilitates the oligomerization of NLRs, which is thought to be a crucial step in their activation. The N-terminal effector domain consists of a protein-protein interaction domain responsible for signaling [2]. NOD2 recognizes muramyl dipeptide (MDP), a component of peptidoglycans from most gram-negative and Gram-positive bacteria, and thus recognizes intracellular bacteria or extracellular bacteria that inject peptidoglycan motifs into the cells [2]. In addition, NOD2 has also been reported to recognize ssRNA [3]. Once activated, NOD2 oligomerizes and interacts with the serine/threonine kinase receptor-interacting protein 2 (RIP2) though the CARD domains [4, 5]. After ubiquitination, RIP2 interacts with transforming growth factor (TGF)-activated kinase 1 (TAK1) [6], resulting in the activation of NF-κB [7] and MAPKs [8], and ultimately increasing the levels of proinflammatory cytokines and the release of antimicrobial peptides [2].
PPRs function to activate both the initial innate immune responses and the subsequent adaptive immune responses. However, their functions must be tightly regulated, as uncontrolled immune responses can damage the host. NOD2 has been reported to be negatively regulated by multiple factors. Caspase 12, an inflammatory cysteine protease, blunts NOD2-induced NF-κB activation by displacing TRAF6 and binding to and ubiquitinating RIP2 [9]. NOD2-S, a truncated form of NOD2, inhibits NOD2/RIP2-induced signaling pathways by interacting with both NOD2 and RIP2 to inhibit the oligomerization of NOD2 [10]. Centaurin β1, a GTPase-activating protein, selectively decreases NF-κB activation induced by NOD1 and NOD2 [11]. A20, a ubiquitin-editing enzyme, playing important roles in regulating TLR signaling [12, 13] and directly restricts NOD2-induced signals in vitro and in vivo [14]. As A20 expression has been reported to be induced by the proinflammatory cytokine IL-1 [15], we postulate that IL-1 negatively regulates NOD2 function. However, in this study, we unexpectedly found that IL-1 pre-treatment neither decreased NOD2-induced production of proinflammatory cytokines nor decreased NOD2-elicited signal transduction in cells, although it induced A20 expression. In contrast, IL-1 promoted MDP-induced cytokine production and signaling by upregulating the expression of NOD2 and its signaling adaptor RIP2.
Materials and methods
Reagents
Rabbit anti-human A20 and GAPDH antibodies, rabbit anti-human phosphorylated ERK, JNK, p38, and NF-κB P65 antibodies, and the ERK inhibitor U0126, were purchased from Cell Signaling Technology (Beverly, MA, USA). The NOD2 ligand MDP was purchased from Invivogen (San Diego, CA, USA). The TLR4 ligand lipopolysaccharide (LPS) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Human IL-8 and IL-1β ELISA kits were purchased from Jiamay Biotech. (Beijing, China). The A20 expression plasmid was purchased from GeneCopoeia (Germantown, MD, USA). The JNK inhibitor SP600125 and P38 inhibitor SB202190 were purchased from Tocris (Bristol, UK). The mouse anti-human NOD2 antibody and NF-κB inhibitor Bay 11–7082 were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Recombinant human IL-1α and IL-1β were purchased from PeproTech Inc. (Rocky Hill, NJ, USA). NF-κB inhibitor BAY 11–7085 was purchased from MedChemExpress (Monmouth Junction, NJ, USA).
Cell culture
THP1, a human monocytic cell line [16], and RAW264.7, a mouse macrophage cell line [17], were purchased from ATCC (Manassas, VA, USA). Cells were cultured in RPMI 1640 containing 10% FCS and antibiotics. All cells were cultured in a humidified atmosphere with 5% CO2 at 37 °C.
Preparation of human peripheral blood mononuclear cells (PBMCs)
Twenty milliliters of fresh whole blood were collected from healthy volunteers in tubes containing the anti-coagulant heparin using standard venipuncture techniques. PBMCs were immediately isolated after blood collection using standard density gradient centrifugation with Ficoll-Paque Plus (Amersham Biosciences AB). Cells were washed and re-suspended in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin, and streptomycin.
Preparation of mouse spleen cells
Female C57BL/6 mice (6–8 weeks old) from the SLAC Laboratory Animal Center (Shanghai, China) were used in all experiments. Animal experiments were approved by the Ethics Committee of Changsha Central Hospital. For the preparation of spleen cells, mice were sacrificed by cervical dislocation. Mouse spleens were removed surgically and minced aseptically. After lysing erythrocytes, spleen cells were washed and cultured in RPMI 1640 supplemented with 10% fetal bovine serum, penicillin, and streptomycin.
Reverse transcription-PCR (RT-PCR)
Total RNA was prepared from 1 to 2 × 106 cells using TRIzol (Invitrogen, Carlsbad, CA, USA), as described by the manufacturer. The mRNA samples were reverse transcribed into cDNAs with RevertAid (MBI Fermentas, Burlington Ontario, Canada) at 42 °C for 60 min, and the resulting cDNAs were subjected to PCR (94 °C for 1 min followed by 20-25 cycles at 94 °C for 30 s, 60 °C for 30 s, and 68 °C for 1 min and an extension for 10 min at 68 °C). PCR products were separated on 1.0% agarose gels and visualized with GelRed (Biotium, Hayward, CA, USA). The following forward and reverse primer pairs (5′–3′) were used:
A20-F: ATGAGGCCAAAAGGACAGAA
A20-R: ACTGAAAGCATTCGTTGCAG
GAPDH-F: AATCCCATCACCATCTTCCA
GAPDH-R: CCTGCTTCACCACCTTCTTG
GRO-α-F: TCACCCCAAGAACATCCAAA
GRO-α-R: TCCTAAGCGATGCTCAAACA
GRO-β-F: GCAGGGAATTCACCTCAAGAA
GRO-β-R: AACACATTAGGCGCAATCCA
IL-1β-F: TTGAAGCTGATGGCCCTAAAC
IL-1β-R: CACCAAGCTTTTTTGCTGTG
IL-6-F: CTTGCCTGGTGAAAATCATC
IL-6-R: TGGACTGCAGGAACTCCTTAA
IL-8-F: TTGGCAGCCTTCCTGATTT
IL-8-R: TCAAAAACTTCTCCACAACCC
mGAPDH-F: CAACTTTGGCATTGTGGAAGG
mGAPDH-R: TCCTCAGTGTAGCCCAAGATG
mGRO-F: TAACCAGTTCCAGCACTCCA
mGRO-R: TTTCTGAACCAAGGGAGCTT
mGRO2-F: TCCAGCCACACTTCAGCCTA
mGRO2-R: TTAGCCTTGCCTTTGTTCAG
mIL-1β-F: GACGGACCCCAAAAGATGAA
mIL-1β-R: CAGCACGAGGCTTTTTTGTT
mIL-6-F: TTGGGACTGATGCTGGTGA
mIL-6-R: ACTCCAGAAGACCAGAGGAAA
mMCP-1-F: TCCTCCACCACCATGCAG
mMCP-1-R: TCTTGGGGTCAGCACAGA
mTNFα-F: CGTCGTAGCAAACCACCAAG
mTNFα-R: GGGGTCAGAGTAAAGGGGTC
NOD2-F: AGCCATGTGGAGAACATGCT
NOD2-R: TGTCCGCATCGTCATTGA
RIP2-R: TGCAGGAAACTCAGAACGTCT
RIP2-R: TATTTCCGGGTAAGGCTGAA
TNFα-F: ATCAGAGGGCCTGTACCTCAT
TNFα-R: AGACTCGGCAAAGTCGAGATA
Immunoblot
Cells (1–2 × 106) were lysed in 200 ml of lysis buffer (20 mM Tris, pH 7.5, 150 mM NaCl, 1% Triton X-100, 1 mM EDTA, 1 mM sodium pyrophosphate, 1 mM β-glycerophosphate, 1 mM Na3VO4, and 1 mg/ml leupeptin). The cell lysate was centrifuged at 12,000 g for 5 min at 4 °C. Proteins were electrophoresed on 10% SDS-PAGE gels and transferred to Immobilon P membranes (Millipore, Billerica, MA, USA). Membranes were blocked with 3% nonfat dry milk for 1 h at room temperature and then incubated with primary antibodies in PBS containing 0.01% Tween 20 overnight at 4 °C. After an incubation with a horseradish peroxidase-conjugated secondary antibody, the protein bands were detected with SuperSignal chemiluminescent substrate-stable peroxide solution (Pierce Rockford, IL, USA) and BIOMAX-MR film (Eastman Kodak Co., Rochester, NY, USA). When necessary, the membranes were stripped with Restore Western blot stripping buffer (Pierce) and re-probed with antibodies against various cellular proteins.
Quantitative RT-PCR (qRT-PCR)
The qRT-PCR assay was performed as described [18, 19]. Briefly, total RNA was isolated and reverse transcribed as described above. The cDNA templates were amplified using TaqMan Universal PCR master mix (Roche Applied Science) and a LightCycle 96 detection system (Roche Applied Science). The amplification of the target genes was normalized to the levels of glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as an endogenous control. The efficiency of PCR was tested by amplifying the target from serially diluted cDNA samples generated from the reverse transcription of a stock set of human RNAs. The data were analyzed and calculations were performed using the 2−ΔΔCT comparative method, as described by the manufacturer. Gene expression is presented as the fold induction of a gene measured in IL-1- and/or MDP-treated samples relative to samples cultured with medium. The same primers were used as described for RT-PCR.
Plasmid transfection
Cells cultured in six-well plates were transfected with 1 μg of plasmid containing the A20 cDNA using LipofectamineTM 2000 (Invitrogen), according to the manufacturer’s instructions. The expression of A20 was measured by western blotting 48 h after transfection. For stable transfection, G418-resistant cells were selected after an incubation with 800 μg/ml G418 for 3 weeks.
Enzyme-linked immunosorbent assay (ELISA)
The levels of IL-1β and IL-8 in culture supernatants were detected using enzyme-linked immunosorbent assays (ELISAs), according to the manufacturer’s standard protocols.
Flow cytometry analysis
Monocytic THP1 cells were treated with IL-1β for 24 h. The cells were harvested and washed with fluorescence-activated cell sorting buffer (5 mmol/L EDTA, 0.1% NaN3, and 1% FCS in Dulbecco’s PBS). After an incubation with a monoclonal antibody against human NOD2 for 30 min on ice, the cells were stained with a FITC-labeled secondary antibody and NOD2 expression was examined by flow cytometry (BD Bioscience, San Jose, CA).
Statistical analysis
All experiments were performed at least three times, and representative results are shown. The results are reported as the mean ± S.D. Differences between groups were examined for statistical significance using Student’s t test, and P values equal to or less than 0.05 were considered statistically significant (n = 3 for each qRT-PCR and ELISA test).
Results
IL-1α/β pre-treatment increases MDP-induced cytokine production in monocytic THP1 cells
As shown in our previous study, IL-1 induces the expression of A20, which in turn regulates IL-1-induced chemokine production in human mesangial cells by inhibiting MAPK signaling [15]. A20 has also been reported to restrict NOD2-triggered signaling in macrophages [14]. Based on these observations, IL-1 possesses the ability to negatively regulate NOD2-induced responses by inducing A20 expression. In this study, we first examined the effect of IL-1 on cytokine production induced by NOD2 activation in monocytic THP1 cells. Unexpectedly, neither IL-1α nor IL-1β downregulated, but upregulated the mRNA levels of proinflammatory cytokines, including IL-1β, IL-6, and TNFα, in cells treated with the NOD2 ligand MDP (Fig. 1a, b). The IL-1 treatment also upregulated the mRNA levels of chemokines, including GROα, β, and IL-8 (Fig. 1c, d). ELISA results showed that IL-1β pre-treatment also increased levels of the IL-1β protein induced by MDP re-stimulation (Fig. 1e). Both IL-1α (Fig. 1f) and IL-1β (Fig. 1g) pre-treatments increased the levels of the IL-8 protein following MDP re-stimulation. Thus, IL-1α/β function to enhance MDP-induced immune responses.
IL-1α/β up-regulate MDP-induced cytokine production in monocytic THP1 cells. a Quantitative RT-PCR analysis of the mRNA levels of proinflammatory cytokines in THP1 cells that were pre-treated with the indicated concentrations of IL-1α for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. b Quantitative RT-PCR analysis of the mRNA levels of proinflammatory cytokines in THP1 cells that were pre-treated with the indicated concentrations of IL-1β for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. c Quantitative RT-PCR analysis of chemokine mRNA levels in THP1 cells that were pre-treated with the indicated concentrations of IL-1α for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. d Quantitative RT-PCR analysis of chemokine mRNA levels in THP1 cells that were pre-treated with the indicated concentrations of IL-1β for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. e ELISA of levels of the IL-1β protein in supernatants from THP1 cells that were pre-treated with the indicated concentrations of IL-1β for 24 h, washed with PBS twice, and re-stimulated with 10 µg/ml MDP for 24 h. f ELISA of levels of the IL-8 protein in the supernatants from THP1 cells that were pre-treated with the indicated concentrations of IL-1α for 24 h and re-stimulated with 10 µg/ml MDP for 24 h. g ELISA of levels of the IL-8 protein in the supernatants from THP1 cells that were pre-treated with the indicated concentrations of IL-1β for 24 h and re-stimulated with 10 µg/ml MDP for 24 h. *P < 0.05 compared with the groups treated with MDP alone
IL-1α/β pre-treatment increases MDP-induced cytokine production in human peripheral blood mononuclear cells (PBMCs), mouse RAW264.7 macrophages, mouse spleen cells, and mouse peritoneal macrophages
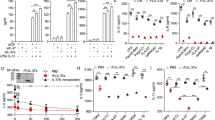
Human PBMCs were pre-treated with IL-1 and re-treated with MDP to determine the effect of IL-1α/β on NOD2-induced inflammation in various immune cells. Cytokine production was detected using quantitative RT-PCR, and the IL-1α and IL-1β pre-treatments both promoted the production of proinflammatory cytokines, including IL-1β, IL-6, and TNFα, in PBMCs stimulated with the NOD2 ligand MDP (Fig. 2a, b). The IL-1α and IL-1β treatment also upregulated the levels of chemokine mRNAs, including GROα, β, and IL-8 (Fig. 2c, d). IL-1α and IL-1β pre-treatments increased the production of IL-1β in mouse RAW264.7 macrophages (Fig. 2e, f). The IL-1β pre-treatment also upregulated MDP-induced IL-6 production in mouse spleen cells (Fig. 2g). Furthermore, the in vivo IL-1β pre-treatment of mouse peritoneal macrophages upregulated MDP-induced production of cytokines, including IL-1β, IL-6, TNFα, MCP-1, GRO, and GRO2 (Fig. 2h). On the basis of these results, IL-1 promotes MDP-induced immune responses in multiple immune cell types.
IL-1α/β up-regulate MDP-induced cytokine production in human peripheral blood mononuclear cells (PBMCs), mouse RAW264.7 macrophages, mouse spleen cells, and mouse peritoneal macrophages. a Quantitative RT-PCR analysis of the mRNA levels of proinflammatory cytokines in PBMCs that were pre-treated with the indicated concentrations of IL-1α for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. b Quantitative RT-PCR analysis of the mRNA levels of proinflammatory cytokines in PBMCs that were pre-treated with the indicated concentrations of IL-1β for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. c Quantitative RT-PCR analysis of chemokine mRNA levels in PBMCs that were pre-treated with the indicated concentrations of IL-1α for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. d Quantitative RT-PCR analysis of chemokine mRNA levels in PBMCs that were pre-treated with the indicated concentrations of IL-1β for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. e Quantitative RT-PCR analysis of the levels of the IL-1β mRNA in mouse RAW264.7 macrophages that were pre-treated with the indicated concentrations of IL-1α for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. f Quantitative RT-PCR analysis of the levels of the IL-1β mRNA in mouse RAW264.7 macrophages that were pre-treated with the indicated concentrations of IL-1β for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. g Quantitative RT-PCR analysis of the levels of the IL-6 mRNA in mouse spleen cells that were pre-treated with the indicated concentrations of IL-1β for 24 h and re-stimulated with 10 µg/ml MDP for 3 h. h C57BL/6 mice were intraperitoneally injected with PBS, or IL-1β (400 ng in 200 μl PBS). 24 h after this injection, the mice were re-injected with MDP (200 μg in 200 μl PBS) intraperitoneally. 6 h after the second injection, mouse peritoneal macrophages were collected, and the cytokine mRNA levels were detected by quantitative RT-PCR. *P < 0.05 compared with the groups treated with MDP alone
The effect of the MDP pre-treatment on IL-1α/β-induced cytokine production
THP1 cells were pre-treated with the NOD2 ligand MDP, re-treated with IL-1α or IL-1β, and cytokine production was measured using quantitative RT-PCR to determine the effect of NOD2 activation on IL-1-induced cytokine production. The MDP pre-treatment exerted a slight effect on IL-1α-induced cytokine production, including a slight inhibition of TNFα (Fig. 3a) and a slight upregulation of GROα and IL-8 (Fig. 3b). The MDP pre-treatment also exerted a slight effect on IL-1β-induced cytokine production, including a slight increase in IL-1β (Fig. 3c) and GROα (Fig. 3d), and a slight inhibition of TNFα (Fig. 3c). Thus, NOD2 has little effect on IL-1α/β-induced inflammatory responses.
The effect of the MDP pre-treatment on IL-1α/β-induced cytokine production in monocytic THP1 cells. a Quantitative RT-PCR analysis of the mRNA levels of proinflammatory cytokines in THP1 cells that were pre-treated with the indicated concentrations of MDP for 24 h and re-stimulated with 20 ng/ml IL-1α for 1 h. b Quantitative RT-PCR analysis of chemokine mRNA levels in THP1 cells that were pre-treated with the indicated concentrations of MDP for 24 h and re-stimulated with 20 ng/ml IL-1α for 1 h. c Quantitative RT-PCR analysis of the mRNA levels of proinflammatory cytokines in THP1 cells that were pre-treated with the indicated concentrations of MDP for 24 h and re-stimulated with 20 ng/ml IL-1β for 1 h. d Quantitative RT-PCR analysis of chemokine mRNA levels in THP1 cells that were pre-treated with the indicated concentrations of MDP for 24 h and re-stimulated with 20 ng/ml IL-1β for 1 h. *P < 0.05 compared with the groups treated with IL-1 alone
IL-1β-induced A20 expression does not regulate NOD2-induced cytokine production
We first detected the expression of A20 in cells stimulated with IL-1β to determine whether A20 is involved in NOD2-induced cytokine production. Both quantitative RT-PCR and western blot results showed that IL-1β upregulated the expression of A20 in a time- (Fig. 4a, b) and dose-dependent manner (Fig. 4c, d). Neither IL-1α nor IL-1β inhibited the expression of ITCH and TAX1BP1 (Fig. 4e, f), two important A20 accessory molecules [20, 21]. Then, A20 was overexpressed in THP1 cells by gene transfer (Fig. 4g, h). When the cells that overexpressed A20 were treated with MDP, quantitative RT-PCR results showed that A20 overexpression did not negatively regulate the MDP-induced expression of cytokines, including GRO-α, IL-8, IL-1β and TNFα (Fig. 4i). As a positive control, A20 overexpression inhibited the LPS-induced production of IL-1β and TNFα (Fig. 4i). Based on these results, IL-1β-induced A20 expression does not regulate MDP-induced immune responses.
A20 does not regulate MDP-induced cytokine production. a Quantitative RT-PCR analysis of A20 mRNA levels in THP1 cells that were treated with 20 ng/ml IL-1β for the indicated time periods. b Western blot analysis of levels of the A20 protein in THP1 cells that were treated with 20 ng/ml IL-1β for the indicated time periods. c Quantitative RT-PCR analysis of A20 mRNA levels in THP1 cells that were treated with the indicated concentrations of IL-1β for 24 h. d Western blot analysis of levels of the A20 protein in THP1 cells that were treated with the indicated concentrations of IL-1β for 24 h. e, f Western blot analysis of levels of the ITCH and TAX1BP1 proteins in THP1 cells that were treated with the indicated concentrations of IL-1α (e) or IL-1β (f) for 24 h. g Quantitative RT-PCR analysis of A20 mRNA levels in THP1 cells that were transfected with the A20 mammalian expression plasmid. h Western blot analysis of levels of the A20 protein in THP1 cells that were transfected with the A20 mammalian expression plasmid. i Quantitative RT-PCR analysis of cytokine mRNA levels in A20-overexpressing THP1 cells that were treated with 10 µg/ml MDP for 3 h. *P < 0.05 compared with the control groups
IL-1α/β up-regulate NOD2 and RIP2 via NF-κB
We measured the effect of IL-1α/β on the expression of NOD2 and its downstream signaling adaptor RIP2 in THP1 cells to explore the mechanism by which IL-1α/β enhanced NOD2-induced immune responses. According to the quantitative RT-PCR results, IL-1β upregulated the levels of the NOD2 and RIP2 mRNAs in a time- (Fig. 5a) and dose-dependent manner (Fig. 5b). FACS results showed that THP1 cells treated with IL-1α and IL-1β exhibited increased levels of the NOD2 protein (Fig. 5c). Western blots revealed high levels of the RIP2 protein in resting THP1 cells, and IL-1α and IL-1β slightly increased the levels of this protein in a dose- (Fig. 5d) and time-dependent manner (Fig. 5e). IL-1α and IL-1β upregulated RIP2 expression in mouse spleen cells (Fig. 5f). As IL-1 receptors activate MAPKs and NF-κB, we tested which pathway contributed to the upregulation of NOD2 and RIP2 induced by IL-1β. Quantitative RT-PCR results showed that NF-κB inhibitor reversed the mRNA upregulation of NOD2 and RIP2 (Fig. 5g). Western blots (Fig. 5h) and FACS (Fig. 5i) revealed that NF-κB inhibitor reversed the protein upregulation of NOD2 and RIP2. Thus, IL-1α/β may promote MDP-induced cytokine production by upregulating NOD2 and RIP2 via NF-κB pathway.
IL-1α/β up-regulates NOD2 and RIP2 via NF-κB pathway. a Quantitative RT-PCR analysis of NOD2 and RIP2 mRNA levels in THP1 cells that were treated with 20 ng/ml IL-1β for the indicated time periods. b Quantitative RT-PCR analysis of NOD2 and RIP2 mRNA levels in THP1 cells that were treated with the indicated concentrations of IL-1β for 3 h. c FACS analysis of levels of the NOD2 protein in THP1 cells that were treated with 20 ng/ml IL-1β or 20 ng/ml IL-1α for 24 h. d Western blot analysis of levels of the RIP2 protein in THP1 cells that were treated with the indicated concentrations of IL-1β or IL-1α for 24 h. e Western blot analysis of levels of the RIP2 protein in THP1 cells that were treated with 20 ng/ml IL-1β or IL-1α for the indicated time periods. f Western blot analysis of levels of the RIP2 protein in mouse spleen cells that were treated with the indicated concentrations of IL-1α for 24 h or with 20 mg/ml IL-1β for the indicated periods. g Quantitative RT-PCR analysis of NOD2 and RIP2 mRNA levels in THP1 cells, pre-treated with the ERK inhibitor U0126 (U, 10 µM), JNK inhibitor SP600125 (SP, 10 µM), P38 inhibitor SB202190 (SB, 10 µM), or NF-κB inhibitor Bay 11-7082 (B82, 10 µM) for 1 h, and re-treated with 20 ng/ml IL-1β for 3 h. h Western blot analysis of the protein levels of RIP2 in THP1 cells, pre-treated with the indicated concentrations of NF-κB inhibitor Bay 11-7082 (B82) for 1 h, and re-treated with 20 ng/ml IL-1β for 24 h. i FACS analysis of levels of the NOD2 protein in THP1 cells, un-treated, or pre-treated with PBS, or 10 µM NF-κB inhibitor Bay 11-7082 (B82) for 1 h, and re-treated with 20 ng/ml IL-1β for 24 h. *P < 0.05 compared with the control groups
IL-1α/β enhance MDP-induced signal transduction
We examined the effect of IL-1α/β on NOD2 signal transduction to further confirm the mechanism by which IL-1 enhances NOD2-induced cytokine production. We first determined the effect of the NOD2 ligand MDP on the phosphorylation of MAPKs and NF-κB p65 in resting THP1 cells. Western blots showed that MDP induced the phosphorylation of MAPKs ERK, JNK, and P38 and the phosphorylation of NF-κB P65 in a time- (Fig. 6a) and dose-dependent (Fig. 6b) manner. THP1 cells were pre-treated with inhibitors of specific signaling pathways and then treated with MDP to confirm which signaling pathway contributed to cytokine production. As evidenced by the quantitative RT-PCR results, ERK contributed to the transcription of the IL-8, GRO-α, TNFα and IL-1β genes, JNK contributed to the transcription of IL-8, and P38 contributed to the transcription of GRO-α (Fig. 6c). As expected, the ELISAs revealed that ERK and P38 pathways contributed to the secretion of the IL-8 protein (Fig. 6d). Unexpectedly, the NF-κB pathway did not contribute to the production of any of the tested cytokines (Fig. 6c, d). When THP1 cells that had been pre-treated with IL-1β were re-treated with MDP, the levels of phosphorylated MAPKs ERK, JNK, P38, and NF-κB P65 were increased, as shown in the western blots (Fig. 6e). Signal transduction was activated in mouse spleen cells pre-treated with IL-1α (Fig. 6f) or IL-1β (Fig. 6g) and re-treated with MDP. To further confirm that NF-κB did not contribute to the upregulation of cytokines induced by MDP, THP1 cells were treated with NF-κB inhibitor Bay 11-7082 or Bay 11-7085 respectively, and then re-treated with MDP. Western blot results showed that both Bay 11-7082 and Bay 11-7085 revered MDP-induced activation of NF-κB (Fig. 6h, i). Quantitative RT-PCR revealed that two NF-κB inhibitors did not reverse MDP-induced cytokine production (Fig. 6j, k). Based on these results, IL-1α/β enhances the MDP-induced immune response by activating NOD2 signal transduction.
IL-1α/β enhance MDP-induced signal transduction. a Western blot analysis of the phosphorylation of the MAPKs ERK, JNK, and P38, as well as NF-κB P65 in THP1 cells that were treated with 10 µg/ml MDP for the indicated time periods. b Western blot analysis of the phosphorylation of the MAPKs ERK, JNK, and P38, as well as NF-κB P65 in THP1 cells that were treated with the indicated concentrations of MDP for 1 h. c Quantitative RT-PCR analysis of cytokine mRNA levels in THP1 cells that were pre-treated with the ERK inhibitor U0126 (U, 10 µM), JNK inhibitor SP600125 (SP, 10 µM), P38 inhibitor SB202190 (SB, 10 µM), or NF-κB inhibitor Bay 11-7082 (Bay, 10 µM) for 1 h, and re-treated with 10 µg/ml MDP for 3 h. d ELISA of levels of the IL-8 protein in the supernatants from THP1 cells that were pre-treated with the ERK inhibitor U0126 (U, 10 µM), JNK inhibitor SP600125 (SP, 10 µM), P38 inhibitor SB202190 (SB, 10 µM), or NF-κB inhibitor Bay 11-7082 (Bay, 10 µM) for 1 h, and re-treated with 10 µg/ml MDP for 24 h. e Western blot analysis of the phosphorylation of the MAPKs ERK, JNK, and P38, as well as NF-κB P65 in THP1 cells that were pre-treated with the indicated concentrations of IL-1β for 24 h and re-treated with 10 µg/ml MDP for 1 h. f Western blot analysis of the phosphorylation of the MAPKs ERK, JNK, and P38, as well as NF-κB P65 in mouse spleen cells that were pre-treated with the indicated concentrations of IL-1α for 24 h and re-treated with 10 µg/ml MDP for 1 h. g Western blot analysis of the phosphorylation of the MAPKs ERK, JNK, and P38, as well as NF-κB P65 in mouse spleen cells that were pre-treated with the indicated concentrations of IL-1β for 24 h and re-treated with 10 µg/ml MDP for 1 h. h, i Western blot analysis of the phosphorylated protein levels of NF-κB P65 in THP1 cells, pre-treated with the indicated concentrations of NF-κB inhibitor Bay 11-7082 (B82) (h), or Bay 11-7085 (B85) (i) for 1 h, and re-treated with 10 µg/ml MDP for 1 h. j, k Quantitative RT-PCR analysis of IL-8 mRNA levels in THP1 cells, pre-treated with the indicated concentrations of NF-κB inhibitor Bay 11-7082 (B82) (h), or Bay 11-7085 (B85) (i) for 1 h, and re-treated with 10 µg/ml MDP for 3 h. *P < 0.05 compared with groups treated with MDP alone
Tolerant cytokine production induced by IL-1β and MDP in THP1 cells
The IL-1α/β pre-treatment increased cytokine production induced by MDP, but the MDP pre-treatment did not inhibit cytokine production induced by IL-1α/β, suggesting that cross-tolerance was not elicited between IL-1 receptor and NOD2. To determine whether the IL-1β or MDP pre-treatment regulates cytokine production induced by the IL-1β or MDP re-stimulation respectively, THP1 cells were pre-treated with MDP for 24 h, re-treated with MDP for 3 h, and the levels of cytokine mRNAs were determined using quantitative RT-PCR. The MDP pre-treatment significantly downregulated the levels of cytokine mRNAs, including IL-1β, IL-6, TNFα, GROα, GROβ, and IL-8 (Fig. 7a, b). When THP1 cells were pre-treated with IL-1β for 24 h and re-treated with IL-1β for 1 h, cytokine production was significantly inhibited as well (Fig. 7c, d). Thus, MDP and IL-1β tolerance was induced in THP1 cells.
IL-1β and MDP induce tolerant cytokine production in THP1 cells. a, b Quantitative RT-PCR analysis of mRNA levels of proinflammatory cytokines (a) and chemokines (b) in THP1 cells that were pre-treated with the indicated concentrations of MDP for 24 h and re-treated with 10 µg/ml MDP for 3 h. c, d Quantitative RT-PCR analysis of mRNA levels of proinflammatory cytokines (c) and chemokines (d) in THP1 cells that were pre-treated with the indicated concentrations of IL-1β for 24 h and re-treated with 20 ng/ml IL-1β for 3 h. *P < 0.05 compared with groups that only received the pre-treatment
Discussion
In this study, we unexpectedly found that the proinflammatory cytokine IL-1 enhanced the MDP-induced immune response. Increased cytokine production depended on ERK, JNK and p38 signaling, but not NF-κB. IL-1 induced the upregulation of the ubiquitin-editing enzyme A20, but it did not regulate MDP-mediated responses. IL-1 increased MDP-induced cytokine production by upregulating NOD2 and RIP2, which contributes to the enhancement of MDP-induced signal transduction.
As shown in our preliminary study, IL-1 induces tolerant production of CXC chemokines in human mesangial cells (HMCs) [15]. The pre-treatment of HMCs with IL-1 decreases cytokine production induced by IL-1 re-stimulation. A20 overexpression inhibits IL-1-induced chemokine production, and A20 downregulation reverses tolerance [15]. On the basis of these results, IL-1-induced A20 expression contributes to the tolerant production of chemokines induced by IL-1. In another study, we found that IL-1β induced A20 expression and subsequently inhibited Pam3CSK4-induced activation of MAPKs [22]. According to the results from our recent study, TNFα-induced A20 production functions to decrease TNFα-induced chemokine production by inhibiting ERK signaling [23]. As A20 is an important regulator of NOD2-induced signal transduction [14], we propose that IL-1 decreases the NOD2-induced immune response by upregulating A20. However, in this study, IL-1 upregulated A20, but the IL-1 pre-treatment did not inhibit and instead increased MDP-induced cytokine production. Additionally, A20 overexpression did not inhibit MDP-induced cytokine production. Thus, IL-1-induced A20 expression does not regulate NOD2-elicited immune responses.
NOD2 is reported to be expressed in all tested mouse tissues, including the spleen, liver, brain, thymus, heart, lung, testicle, ovary, kidney, and embryo, with high levels detected in the spleen, thymus, and lung [24]. In humans, NOD2 is expressed by multiple cell types, such as neutrophils [25], macrophages [26], monocytes [27], eosinophils [28], basophils [28], B lymphocytes [29], T lymphocytes [30], mast cells [31], dendritic cells [32], stem cells [33], Paneth cells [34], and intestinal epithelial cells [35]. NOD2 expression is regulated by numerous factors. TNFα alone or in combination with IFNγ up-regulates NOD2 expression in epithelial cells [24, 36]. The NOD2 level increases in response to lipopolysaccharide and lipoteichoic acid [24, 37]. Resistin, a type of adipokine, has been reported to upregulate NOD2 expression in mouse monocytes [37]. Levels of the NOD2 protein are increased in response to Aspergillus fumigatus conidia both in vitro and in vivo [38]. In addition, NOD2 expression has been reported to be downregulated by several factors, such as ghrelin [39], cigarette smoke extract [40], baicalin [41], and microRNAs such as miR-192, miR-495, miR-512, and miR-671 [42]. However, the effects of the proinflammatory cytokine IL-1 on NOD2 expression and functions are not completely understood. In the present study, IL-1 upregulated the expression of NOD2 and its downstream adaptor RIP2 at both the mRNA and protein levels, resulting in increased MDP-induced signal transduction and ultimately an increase in cytokine production induced by NOD2 activation.
NOD2 recognizes MDP and triggers downstream signaling pathways, including activation of NF-κB [43, 44] and MAPKs [45, 46], which leads to the activation of inflammatory immune responses. NOD2 has been reported to induce TLR2-mediated IL-23 expression via NF-κB in Paneth cell-like cells [47]. Z-100, an extract from Mycobacterium tuberculosis, increases the production of the proinflammatory cytokine TNFα via a NOD2-dependent NF-κB signaling pathway [44]. By activating NF-κB pathways, NOD2 has been reported to induce the production of the CC chemokine CCL5 [48]. Therefore, the inhibition of NOD2-mediated NF-κB activation reduces the inflammatory response [49]. NOD2 has been reported to induce IL-8 expression via an NF-κB-independent pathway [45]. In addition, NOD2 exerts its non-inflammatory functions via NF-κB-independent pathways. Following the activation of ERK signaling in subjects with diabetic nephropathy, NOD2 increases the endothelial-to-mesenchymal transition of glomerular endothelial cells [50]. Both NOD1 and NOD2 have been reported to control the invasiveness of trophoblast cells via the MAPK/P38 signaling pathway [45]. In the present study, the activation of NOD2 induced cytokine production via the ERK, JNK, and P38 pathways, but not the NF-κB pathway. IL-1 promoted NOD2-induced cytokine production by increasing NOD2-induced signal transduction through ERK, JNK, and P38.
The association of NOD2 mutations with a number of inflammatory pathologies, including Crohn’s disease (CD) [51], graft-versus-host disease (GVHD) [52, 53], and Blau syndrome [54], highlights its pivotal role in host-pathogen interactions and inflammatory responses. NOD2 has also been reported to be associated with cardiovascular diseases. In a Caucasian population, NOD2/CARD15 polymorphisms influenced the development of clinically evident and angiographically documented coronary artery disease [55]. NOD2-mediated innate immune signaling regulates the eicosanoid levels in subjects with atherosclerosis [56]. NOD2 deletion promotes cardiac hypertrophy and fibrosis induced by pressure overload [57]. Furthermore, NOD2 has been reported to be associated with ischemia/reperfusion injury by regulating apoptosis and inflammation [41, 58, 59]. Downregulation of NOD2 decreases injury [38, 41, 60]. In the present study, IL-1 induced the expression of NOD2 and RIP2, and increased cytokine production, indicating that IL-1 might enhance the injury induced by NOD2 activation.
The results presented in this study are compatible with the model outlined in Fig. 8. IL-1R1, expressed on the cell surface, is activated by IL-1α and IL-1β, resulting in the activation of signal pathways, including MAPKs (ERK, JNK, and P38) and NF-κB. NF-κB activation contributes to the upregulation of NOD2 and RIP2, leading to the upregulation of the signal transduction elicited by MDP activation of NOD2. Thus, IL-1α and IL-1β enhance the cytokine production induced by NOD2.
Model of the pathways by which IL-1α/β up-regulate MDP-induced cytokines. IL-1α and IL-1β activate IL-1R1, resulting in the upregulation of intracellular MDP receptor NOD2 and its signaling adaptor RIP2 via NF-κB pathway. The upregulation of NOD2 and RIP2 increases the activation of ERK and P38 induced by MDP, and thus promotes the production of cytokines
References
Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801.
Coll RC, O’Neill LA. New insights into the regulation of signalling by toll-like receptors and nod-like receptors. J Innate Immun. 2010;2:406–21.
Sabbah A, Chang TH, Harnack R, et al. Activation of innate immune antiviral responses by Nod2. Nat Immunol. 2009;10:1073–80.
Park JH, Kim YG, McDonald C, et al. RICK/RIP2 mediates innate immune responses induced through Nod1 and Nod2 but not TLRs. J Immunol. 2007;178:2380–6.
Fridh V, Rittinger K. The tandem CARDs of NOD2: intramolecular interactions and recognition of RIP2. PLoS ONE. 2012;7:e34375.
Sicard P, Jacquet S, Kobayashi KS, et al. Pharmacological postconditioning effect of muramyl dipeptide is mediated through RIP2 and TAK1. Cardiovasc Res. 2009;83:277–84.
Hasegawa M, Fujimoto Y, Lucas PC, et al. A critical role of RICK/RIP2 polyubiquitination in Nod-induced NF-kappaB activation. EMBO J. 2008;27:373–83.
Tao M, Scacheri PC, Marinis JM, et al. ITCH K63-ubiquitinates the NOD2 binding protein, RIP2, to influence inflammatory signaling pathways. Curr Biol. 2009;19:1255–63.
LeBlanc PM, Yeretssian G, Rutherford N, et al. Caspase-12 modulates NOD signaling and regulates antimicrobial peptide production and mucosal immunity. Cell Host Microbe. 2008;3:146–57.
Rosenstiel P, Huse K, Till A, et al. A short isoform of NOD2/CARD15, NOD2-S, is an endogenous inhibitor of NOD2/receptor-interacting protein kinase 2-induced signaling pathways. Proc Natl Acad Sci USA. 2006;103:3280–5.
Yamamoto-Furusho JK, Barnich N, Xavier R, et al. Centaurin beta1 down-regulates nucleotide-binding oligomerization domains 1- and 2-dependent NF-kappaB activation. J Biol Chem. 2006;281:36060–70.
Catrysse L, Vereecke L, Beyaert R, et al. A20 in inflammation and autoimmunity. Trends Immunol. 2014;35:22–31.
Ma A, Malynn BA. A20: linking a complex regulator of ubiquitylation to immunity and human disease. Nat Rev Immunol. 2012;12:774–85.
Hitotsumatsu O, Ahmad RC, Tavares R, et al. The ubiquitin-editing enzyme A20 restricts nucleotide-binding oligomerization domain containing 2-triggered signals. Immunity. 2008;28:381–90.
Luo H, Liu Y, Li Q, et al. A20 regulates IL-1-induced tolerant production of CXC chemokines in human mesangial cells via inhibition of MAPK signaling. Sci Rep. 2015;5:18007.
Liu B, Sun R, Luo H, et al. Both intrinsic and extrinsic apoptotic pathways are involved in Toll-like receptor 4 (TLR4)-induced cell death in monocytic THP-1 cells. Immunobiology. 2017;222:198–205.
Huang C, Liu XJ. QunZhou, et al. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264.7 macrophages. Int Immunopharmacol. 2016;32:46–54.
Sun R, Zhang Y, Lv Q, et al. Toll-like receptor 3 (TLR3) induces apoptosis via death receptors and mitochondria by up-regulating the transactivating p63 isoform alpha (TAP63alpha). J Biol Chem. 2011;286:15918–28.
Sun Y, Zhu D, Wang G, et al. Pro-inflammatory cytokine IL-1beta up-regulates CXC chemokine receptor 4 via Notch and ERK signaling pathways in tongue squamous cell carcinoma. PLoS One. 2015;10:e0132677.
Shembade N, Harhaj NS, Parvatiyar K, et al. The E3 ligase Itch negatively regulates inflammatory signaling pathways by controlling the function of the ubiquitin-editing enzyme A20. Nat Immunol. 2008;9:254–62.
Parvatiyar K, Barber GN, et al. TAX1BP1 and A20 inhibit antiviral signaling by targeting TBK1-IKKi kinases. J Biol Chem. 2010;285:14999–5009.
Hu J, Wang G, Liu X, et al. A20 is critical for the induction of Pam3CSK4-tolerance in monocytic THP-1 cells. PLoS ONE. 2014;9:e87528.
Zhou X, An D, Liu X, et al. TNFalpha induces tolerant production of CXC chemokines in colorectal cancer HCT116 cells via A20 inhibition of ERK signaling. Int Immunopharmacol. 2018;54:296–302.
Iwanaga Y, Davey MP, Martin TM, et al. Cloning, sequencing and expression analysis of the mouse NOD2/CARD15 gene. Inflamm Res. 2003;52:272–6.
Oh SJ, Kim JH, Chung DH. NOD2-mediated suppression of CD55 on neutrophils enhances C5a generation during polymicrobial sepsis. PLoS Pathog. 2013;9:e1003351.
Juarez E, Carranza C, Hernandez-Sanchez F, et al. NOD2 enhances the innate response of alveolar macrophages to Mycobacterium tuberculosis in humans. Eur J Immunol. 2012;42:880–9.
Beynon V, Cotofana S, Brand S, et al. NOD2/CARD15 genotype influences MDP-induced cytokine release and basal IL-12p40 levels in primary isolated peripheral blood monocytes. Inflamm Bowel Dis. 2008;14:1033–40.
Jiao D, Wong CK, Qiu HN, et al. NOD2 and TLR2 ligands trigger the activation of basophils and eosinophils by interacting with dermal fibroblasts in atopic dermatitis-like skin inflammation. Cell Mol Immunol. 2016;13:535–50.
Lin Z, Hegarty JP, John G, et al. NOD2 mutations affect muramyl dipeptide stimulation of human B lymphocytes and interact with other IBD-associated genes. Dig Dis Sci. 2013;58:2599–607.
Dong W, Zhang H, Yin X, et al. Oral delivery of tumor microparticle vaccines activates NOD2 signaling pathway in ileac epithelium rendering potent antitumor T cell immunity. Oncoimmunology. 2017;6:e1282589.
Okumura S, Yuki K, Kobayashi R, et al. Hyperexpression of NOD2 in intestinal mast cells of Crohn’s disease patients: preferential expression of inflammatory cell-recruiting molecules via NOD2 in mast cells. Clin Immunol. 2009;130:175–85.
Lemire P, Roy D, Fittipaldi N, et al. Implication of TLR- but not of NOD2-signaling pathways in dendritic cell activation by group B Streptococcus serotypes III and V. PLoS ONE. 2014;9:e113940.
Nigro G, Rossi R, Commere PH, et al. The cytosolic bacterial peptidoglycan sensor Nod2 affords stem cell protection and links microbes to gut epithelial regeneration. Cell Host Microbe. 2014;15:792–8.
Rocha JD, Schlossmacher MG, Philpott DJ. LRRK2 and Nod2 promote lysozyme sorting in Paneth cells. Nat Immunol. 2015;16:898–900.
Negroni A, Colantoni E, Vitali R, et al. NOD2 induces autophagy to control AIEC bacteria infectiveness in intestinal epithelial cells. Inflamm Res. 2016;65:803–13.
Rosenstiel P, Fantini M, Brautigam K, et al. TNF-alpha and IFN-gamma regulate the expression of the NOD2 (CARD15) gene in human intestinal epithelial cells. Gastroenterology. 2003;124:1001–9.
Keller JF, Carrouel F, Staquet MJ, et al. Expression of NOD2 is increased in inflamed human dental pulps and lipoteichoic acid-stimulated odontoblast-like cells. Innate Immun. 2011;17:29–34.
Zhang H, Zhu T, Liu W, et al. TIPE2 acts as a negative regulator linking NOD2 and inflammatory responses in myocardial ischemia/reperfusion injury. J Mol Med (Berl). 2015;93:1033–43.
Peng Z, Zhu Y, Zhang Y, et al. Effects of ghrelin on pulmonary NOD2 mRNA expression and NF-kappaB activation when protects against acute lung injury in rats challenged with cecal ligation and puncture. Int Immunopharmacol. 2012;13:440–5.
Aldhous MC, Soo K, Stark LA, et al. Cigarette smoke extract (CSE) delays NOD2 expression and affects NOD2/RIPK2 interactions in intestinal epithelial cells. PLoS ONE. 2011;6:e24715.
Li H, Hu J, Ma L, et al. Comprehensive study of baicalin down-regulating NOD2 receptor expression of neurons with oxygen-glucose deprivation in vitro and cerebral ischemia-reperfusion in vivo. Eur J Pharmacol. 2010;649:92–99.
Chuang AY, Chuang JC, Zhai Z, et al. NOD2 expression is regulated by microRNAs in colonic epithelial HCT116 cells. Inflamm Bowel Dis. 2014;20:126–35.
Hu C, Sun L, Hu Y, Lu D, et al. Functional characterization of the NF-kappaB binding site in the human NOD2 promoter. Cell Mol Immunol. 2010;7:288–95.
Katsunuma K, Yoshinaga K, Ohira Y, et al. Z-100, extracted from Mycobacterium tuberculosis strain Aoyama B, promotes TNF-alpha production via nucleotide-binding oligomerization domain containing 2 (Nod2)-dependent NF-kappaB activation in RAW264.7 cells. Mol Immunol. 2015;64:218–27.
Wang Z, Liu M, Nie X, et al. NOD1 and NOD2 control the invasiveness of trophoblast cells via the MAPK/p38 signaling pathway in human first-trimester pregnancy. Placenta. 2015;36:652–60.
Hedl M, Abraham C. Nod2-induced autocrine interleukin-1 alters signaling by ERK and p38 to differentially regulate secretion of inflammatory cytokines. Gastroenterology. 2012;143:1530–43.
Tan G, Liang E, Liao K, et al. NOD2 up-regulates TLR2-mediated IL-23p19 expression via NF-kappaB subunit c-Rel in Paneth cell-like cells. Oncotarget. 2016;7:63651–60.
Werts C, le Bourhis L, Liu J, et al. Nod1 and Nod2 induce CCL5/RANTES through the NF-kappaB pathway. Eur J Immunol. 2007;37:2499–508.
Zhao H, Li S, Zhang H, et al. Saikosaponin A protects against experimental sepsis via inhibition of NOD2-mediated NF-kappaB activation. Exp Ther Med. 2015;10:823–7.
Shang J, Zhang Y, Jiang Y, et al. NOD2 promotes endothelial-to-mesenchymal transition of glomerular endothelial cells via MEK/ERK signaling pathway in diabetic nephropathy. Biochem Biophys Res Commun. 2017;484:435–41.
Sidiq T, Yoshihama S, Downs I, et al. Nod2: a critical regulator of ileal microbiota and Crohn’s Disease. Front Immunol. 2016;7:367.
Penack O, Smith OM, Cunningham-Bussel A, et al. NOD2 regulates hematopoietic cell function during graft-versus-host disease. J Exp Med. 2009;206:2101–10.
van der Velden WJ, Blijlevens NM, Maas FM, et al. NOD2 polymorphisms predict severe acute graft-versus-host and treatment-related mortality in T-cell-depleted haematopoietic stem cell transplantation. Bone Marrow Transplant. 2009;44:243–8.
Dugan J, Griffiths E, Snow P, et al. Blau syndrome-associated Nod2 mutation alters expression of full-length NOD2 and limits responses to muramyl dipeptide in knock-in mice. J Immunol. 2015;194:349–57.
Galluzzo S, Patti G, Dicuonzo G, et al. Association between NOD2/CARD15 polymorphisms and coronary artery disease: a case-control study. Hum Immunol. 2011;72:636–40.
Liu HQ, Zhang XY, Edfeldt K, et al. NOD2-mediated innate immune signaling regulates the eicosanoids in atherosclerosis. Arterioscler Thromb Vasc Biol. 2013;33:2193–201.
Zong J, Salim M, Zhou H, et al. NOD2 deletion promotes cardiac hypertrophy and fibrosis induced by pressure overload. Lab Invest. 2013;93:1128–36.
Liu H, Wei X, Kong L, et al. NOD2 is involved in the inflammatory response after cerebral ischemia-reperfusion injury and triggers NADPH oxidase 2-derived reactive oxygen species. Int J Biol Sci. 2015;11:525–35.
Liu Y, Yang H, Liu LX, et al. NOD2 contributes to myocardial ischemia/reperfusion injury by regulating cardiomyocyte apoptosis and inflammation. Life Sci. 2016;149:10–17.
Bai X, Zhang X, Chen L, et al. Protective effect of naringenin in experimental ischemic stroke: down-regulated NOD2, RIP2, NF-kappaB, MMP-9 and up-regulated claudin-5 expression. Neurochem Res. 2014;39:1405–15.
Acknowledgements
This study was supported by a grant from the National Science and Technology Major Project of China (2013ZX10005004).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Li, S., Deng, P., Wang, M. et al. IL-1α and IL-1β promote NOD2-induced immune responses by enhancing MAPK signaling. Lab Invest 99, 1321–1334 (2019). https://doi.org/10.1038/s41374-019-0252-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-019-0252-7
This article is cited by
-
IL-1β promotes A7r5 and HASMC migration and invasion via the p38-MAPK/Angpt-2 pathway
European Journal of Medical Research (2022)
-
IL-1 and autoinflammatory disease: biology, pathogenesis and therapeutic targeting
Nature Reviews Rheumatology (2022)
-
NOD2 is involved in regulating odontogenic differentiation of DPSCs suppressed by MDP through NF-κB/p65 signaling
Cytotechnology (2022)
-
Curcumin-primed human BMSC-derived extracellular vesicles reverse IL-1β-induced catabolic responses of OA chondrocytes by upregulating miR-126-3p
Stem Cell Research & Therapy (2021)
-
Downregulation of NUDT21 contributes to cervical cancer progression through alternative polyadenylation
Oncogene (2021)