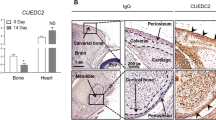
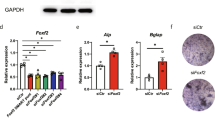
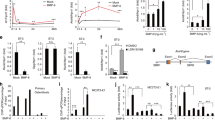
Abstract
Osteoblast differentiation plays a critical role in bone formation and maintaining balance in bone remodeling. Runt-related transcription factor 2 (Runx2) is a central transcription factor regulating osteoblast differentiation and promoting bone mineralization. Until now, the molecular regulatory basis and especially the gene regulatory network of osteogenic differentiation have been unclear. Krüppel-like factor 2 (KLF2) is a zinc finger structure and DNA-binding transcription factor. The current study aimed to investigate the physiological function of KLF2 in osteoblast differentiation. Our results indicate that KLF2 is expressed in pre-osteoblast MC3T3-E1 cells and primary osteoblasts. Interestingly, KLF2 expression is increased in osteoblasts during the osteoblastic differentiation process. Overexpression of KLF2 in MC3T3-E1 cells promoted the expression of the osteoblastic differentiation marker genes Alp, Osx, and Ocn, and stimulated mineralization by increasing Runx2 expression at both the mRNA and protein levels. In contrast, knockdown of KLF2 produced the opposite effects. Importantly, we found that KLF2 could physically interact with Runx2. KLF2 promoted osteoblast differentiation by regulating Runx2 and physically interacting with Runx2. Taken together, the findings of this study identify KLF2 as a novel regulator of osteoblast differentiation. Our findings suggest that KLF2 might be a new therapeutic target for bone disease.
Similar content being viewed by others
Introduction
Bone is a dynamic tissue that can replace old or impaired tissue through the process of bone remodelling [1]. Bone turnover is mainly regulated by osteoblasts and osteoclasts. Osteoblasts are responsible for bone formation, while osteoclasts are responsible for bone resorption [2]. Bone remodeling imbalance has been found in several bone metabolic diseases, such as osteoporosis [3]. Osteoblast differentiation and bone formation play a pivotal role in maintaining the bone remodeling balance and bone mass [4]. The regulation of osteoblast differentiation is complex. Various extracellular and intracellular molecules such as transcriptional factors and posttranslational modifiers have been shown to be involved. Members of the Runt-related transcription factor (Runx) family regulate the expression of genes involved in cellular differentiation and cell cycle progression [5]. Runx2 is an important member of the Runx family and a central transcription factor that induces the commitment of mesenchymal stem cells to osteogenic lineage. Runx2 also participates in bone mineralization by stimulating osteoblast differentiation [6]. The underlying mechanism whereby Runx2 regulates osteoblast differentiation is complicated. A variety of post-translational modifications including methylation, acetylation, and phosphorylation have been reported to regulate the activation of Runx2. Runx2 concomitantly interacts with other factors to initiate the expression of osteoblast differentiation marker genes, such as alkaline phosphatase (Alp), osterix (Osx), and osteocalcin (Ocn) [7]. However, the underlying molecular networks involved in this process have yet to be elucidated.
Krüppel-like factor 2 (KLF2) is one of the most important members of Krüppel-like factor (KLF) family. KLF2 is a zinc finger structure and DNA-binding transcription factor [8]. Previous studies have shown that KLF2 possesses multiple biological functions. It has been reported that KLF2 regulates cellular growth and differentiation [9]. KLF2 has been reported to regulate endothelial functions by preventing laminar shear stress and inhibiting pro-inflammatory cytokine productions. In addition, KLF2 can suppress the expression of vascular cell adhesion molecule-1 (VCAM-1) in endothelial cells by interacting with NF-κB [10]. KLF2 has also been identified as a key regulator of the blood brain barrier (BBB) and a novel neuroprotective factor in ischemic stroke [11]. Interestingly, KLF2 is expressed in primary chondrocytes and can prevent the degradation of type II collagen, a primary component of the extracellular matrix in articular cartilage, by inhibiting the expression of matrix metalloproteinase-13 (MMP-13) [12]. However, the physiological function of KLF2 on osteoblast differentiation and bone formation has not been reported before. The current study was designed to explore the mechanisms through which KLF2 might impact osteoblast differentiation by targeting Runx2.
Materials and methods
Cell culture, treatment, and transfection
Murine pre-osteoblastic MC3T3-E1 cells and human umbilical vein endothelial cells (HUVECs) were purchased from the American Tissue Culture Collection (ATCC, USA). Cells were cultured in a humidified atmosphere containing 5% CO2 at 37 °C. MC3T3-E1 cells were maintained in α-minimal essential medium (α-MEM) (Life Technologies, USA) supplemented with 10% fetal bovine serum (FBS) (Life Technologies, USA). HUVECs were maintained in endothelial cell growth medium BulletKit package (Lonza, USA) containing basal medium, growth factors, cytokines, and supplements. Primary murine calvarial osteoblasts were isolated from the calvaria of neonatal C57BL/6 mice by sequential collagenase digestion following the previous protocols [13]. All animal experiments were performed under a protocol approved by the Institutional Animal Care and Use Committee at the First Affiliated Hospital of Soochow University and were in accordance with the First Affiliated Hospital of Soochow University guidelines for the care and use of laboratory animals. To knock down the expression of KLF2, MC3T3-E1 cells were transfected with KLF2 siRNA using Lipofectamine 2000 (Life Technologies, USA) according to the manufacturer’s protocols. To overexpress KLF2, MC3T3-E1 cells were transduced with lentivirus encoding for KLF2. Osteogenic medium (OM) (10% FBS, 50 μg/mL ascorbic acid, 5 mM β-glycopyrophosphate, and 100 μg/mL BMP2) was used to induce osteoblast differentiation. Culture medium was replaced every 3 days.
HEK-293 T cells were maintained in DMEM supplemented with 10% FBS (Life Technologies, USA) and 1% antibiotics. For immunoprecipitation experiments, HEK-293 T cells were transfected with Flag-tagged KLF2 or Myc-tagged Runx2 using Lipofectamine 2000 (Life Technologies, USA) according to the manufacturer’s protocols.
Isolation of bone marrow mesenchymal stem cells
Sixteen rats were used in this study from which bone marrow, gingival, and submandibular salivary gland samples were collected. Bone marrow cells were isolated from tibia and fibula with PBS containing 2 mM EDTA. Samples were centrifuged for 35 min at 400 × g rpm. The mononuclear cell (MNC) layer at the interphase was collected and centrifuged for 10 min at 200 × g rpm at 10 °C. After two gentle washes with PBS, the cell pellet was re-suspended in a final volume of 300 μl of buffer. Isolated MSCs were maintained in minimal essential medium (MEM) supplemented with 15% FBS at 37 °C and 5% CO2.
Reverse-transcription polymerase chain reaction (PCR)
After the necessary treatment and transfection, total intracellular RNA was isolated from each sample using Trizol reagent (Life Technologies, USA) following the manufacturer’s instructions. One icrogram purified RNA was used for cDNA synthesis using OligodT primer and the iScript cDNA synthesis kit (Bio-Rad, USA). Two microliters of synthesized cDNA was used for standard PCR with dNTPs mix and DNA polymerase. Five microliters of the PCR products was loaded onto 1.5% agarose gel electrophoresis with ethidium bromide (EB).
Quantitative real-time polymerase chain reaction (PCR)
After the necessary treatment and transfection, total intracellular RNA was isolated from each sample using Trizol reagent (Life Technologies, USA) following the manufacturer’s instructions. One microgram purified RNA was used for cDNA synthesis using OligodT primer and the iScript cDNA synthesis kit (Bio-Rad, USA). Synthesized cDNA was used for real-time PCR analysis with SYBR Green master mix (Bio-Rad, USA) on an ABI 7500 real-time PCR system (Applied Biosystems, USA) according to the manufacturer’s instructions. The mRNA value for the target gene was normalized relative to the GAPDH mRNA level in the RNA sample by the 2−∆∆CT method.
Luciferase assay
Briefly, Alp, Osx, and Ocn gene promoter-luciferase reporters, flag-tagged KLF2, Myc-tagged mouse Runx2, and pcDNA3.1 (as an empty vector) plasmids were transfected into MC3T3-E1 cells using Lipofectamine 2000 reagents (Life Technologies, USA) and incubated for 24 h. β-galactosidase plasmid was used as an internal control. Cells were lysed and luciferase activity was measured using a luciferase reporter assay system (Promega, USA) on a multi-plate reader (Bio-Tek Instruments, USA).
Alizarin red staining
An alizarin red staining assay was performed to evaluate mineralization. Briefly, MC3T3-E1 cells were fixed with 70% ice-cold ethanol for 1 h. After washing three times with PBS, cells were incubated with 20 mM of alizarin red solution for 15 min at room temperature (RT). After washing with PBS three times, stained cells were photographed. Then, 10% cetylpyridinium chloride was added to extract the stained cells. Absorbance was recorded at 562 nm using a spectrophotometer (NanoDrop Technologies, USA) and analyzed using Image J software (National Institutes of Health).
Immunoprecipitation
Briefly, HEK-293 T cells or MC3T3-E1 cells were collected using cell lysis buffer (Cell Signaling Technologies, USA) containing protease inhibitor cocktail (Sigma-Aldrich, USA). Protein lysates were subjected to centrifugation at 12,000 × g for 15 min at 4 °C. Supernatants were collected and pre-cleared with protein G–agarose beads (Life Technologies, USA), followed by incubation with anti-Flag (Abcam, USA) and anti-Myc antibodies (Thermo Fisher Scientific) overnight at 4 °C. After incubation with protein G–agarose beads for 4 h, cell lysates were washed and used for western blot analysis.
Western blot analysis
Total intracellular proteins were extracted from the cells with lysis buffer (Cell Signaling Technologies, USA). Protein concentration was determined using a bicinchoninic acid (BCA) assay kit (Beyotime, China). Equal amounts (20 μg) of protein from each sample were separated on 10% sodium dodecyl sulfate polyacrylamide gel (SDS-PAGE) and electrotransferred onto polyvinylidene difluoride (PVDF) membranes. Membranes were blocked with 5% fat-free dry milk in 0.1 M Tris-buffered saline-0.1% Tween-20 buffer (TBST) for 2 h. Then, the membranes were sequentially incubated with the primary antibodies and horseradish peroxidase-conjugated secondary antibody for 1 h. Blots were developed with an enhanced chemiluminescence reagent (Amersham Biosciences, USA) and quantified by densitometric scanning and analyses using an ImageQuant LAS 4000 system (Fujifilm, Tokyo, Japan). β-actin protein was used as an internal control.
Determination of ALP activity
MC3T3-E1 cells were transfected with KLF2 siRNA or transduced with lentivirus encoding for KLF2, followed by stimulation with OM for 14 days. Cells were then assayed for ALP activity with a p-nitrophenyl phosphate (pNPP) liquid substrate system (Sigma) following the manufacturer’s instructions. OD value was recorded at 405 nm on a multifunction microplate scanner (Infinite M200, TECAN).
Statistical analysis
Experimental data are expressed as means ± standard error of mean (S.E.M). Statistical comparisons were carried out by one-way or two-way analysis of variance (ANOVA) followed by post hoc analysis via Scheffe test using SPSS software (version 20.0 USA). P value less than 0.05 was considered statistical significant.
Results
Firstly, we investigated the expression patterns of KLF2 in osteoblasts. As shown in Fig. 1a, RT-PCR analysis results indicate that KLF2 was expressed in primary calvarial osteoblasts and pre-osteoblast MC3T3-E1 cells at the mRNA level. HUVECs were used as a positive control. In addition, the western blot results in Fig. 1b indicate that KLF2 was expressed in the primary calvarial osteoblasts and pre-osteoblast MC3T3-E1 cells. To investigate the influence of KLF2 on osteoblast differentiation, we measured KLF2 expression in the osteoblastic differentiation process of pre-osteoblast MC3T3-E1 cells. Interestingly, we found that KLF2 expression was significantly increased in a time-dependent manner from 3 to 9 days in response to OM treatment at both the mRNA level (Fig. 2a) and protein level (Fig. 2b). Additionally, our results also indicate that stimulation with OM promoted the expression of osteoblast marker genes, such Alp, Osx, and Ocn in a time-dependent manner at both the mRNA level (Fig. 2c) and the protein level (Fig. 2d). The transcriptional factor Runx2 is a central regulator of osteoblast differentiation, which controls the transcription of Alp, Osx, and Ocn. Importantly, our results displayed that stimulation with OM promoted the expression of Runx2 in a time-dependent manner at both the mRNA (supplementary Fig. 1A) and protein levels (supplementary Fig. 1B). To further confirm the effects of KLF2 on osteoblastic differentiation, primary calvarial osteoblasts were used. As expected, stimulation with OM promoted the expression of KLF2 (Fig. 3a) along with an increase in Alp, Osx, and Ocn (Fig. 3c) at the mRNA level. Consistently, we found that western blot analysis revealed an increase in expression of KLF2 (Fig. 3b), Alp, Osx, and Ocn (Fig. 3d) in primary calvarial osteoblasts at the protein level. These results implicate that KLF2 might be associated with osteoblast differentiation.
Krüppel-like factor-2 (KLF2) is expressed in pre-osteoblast MC3T3-E1 cells and primary calvarial osteoblasts. Human umbilical vein endothelial cells (HUVECs) were used as a positive control. a Expression of KLF2 at the mRNA level was determined by reverse-transcription PCR (RT-PCR) analysis; b Expression of KLF2 at the protein level was determined by western blot analysis
Krüppel-like factor-2 (KLF2) expression is increased during osteoblast differentiation in pre-osteoblast MC3T3-E1 cells. Pre-osteoblast MC3T3-E1 cells were cultured in osteogenic medium (OM) for 0, 3, 6, and 9 days. a Expression of KLF2 at the mRNA level was determined by real-time PCR analysis; b Expression of KLF2 at the protein level was determined by western blot analysis; c Expression of alkaline phosphatase (Alp), osterix (Osx), and osteocalcin (Ocn) at the mRNA levels was determined by real-time PCR analysis; d Expression of alkaline phosphatase (Alp), osterix (Osx), and osteocalcin (Ocn) at the protein level was determined by western blot analysis (*, #, $, P < 0.01 vs. previous column group)
Krüppel-like factor-2 (KLF2) expression is increased during osteoblast differentiation in primary calvarial osteoblasts. Primary calvarial osteoblasts were cultured in osteogenic medium (OM) for 0, 3, 6, and 9 days. a Expression of KLF2 at the mRNA level was determined by real-time PCR analysis; b Expression of KLF2 at the protein level was determined by western blot analysis; c Expression of alkaline phosphatase (Alp), osterix (Osx), and osteocalcin (Ocn) at the mRNA level was determined by real-time PCR analysis; d Expression of alkaline phosphatase (Alp), osterix (Osx), and osteocalcin (Ocn) at the protein level was determined by western blot analysis (*, #, $, P < 0.01 vs. previous column group)
To ascertain the effects of KLF2 on osteoblast differentiation, MC3T3-E1 cells were transfected with KLF2 siRNA to knock down the expression of KLF2 and then incubated with OM for 6 days. Successful knockdown of KLF2 is shown in Fig. 4a. As expected, knockdown of KLF2 significantly abolished OM stimulation-induced expression of Alp, Osx, and Ocn at both the mRNA level (Fig. 4b) and the protein level (Fig. 4c). Bone mineralization has been extensively used to characterize osteoblast differentiation [14]. Alizarin Red S staining assay was used to determine the effects of KLF2 on mineralization in MC3T3-E1 cells. The results in Fig. 4d indicate that knockdown of KLF2 reduced OM-induced mineralization of MC3T3-E1 cells. MC3T3-E1 cells were transfected with KLF2 siRNA, followed by stimulation with OM for 14 days. ALP activity in MC3T3-E1 cells was then examined to evaluate osteoinductive activity. The results indicate that silencing of KLF2 reduced the OM-induced increase in ALP activity (Fig. 4e).
Knockdown of KLF2 prevents osteoblast differentiation. MC3T3-E1 cells were transfected with KLF2-specific small RNA interference or non-specific small RNA interference. NS, non-specific small RNA interference; KLF2 siRNA, KLF2 specific small RNA interference. a Western blot analysis revealed the successful knockdown of KLF2; b Real-time PCR analysis revealed that silencing of KLF2 significantly abolished OM stimulation-induced expression of Alp, Osx, and Ocn at the mRNA level; c Western blot analysis revealed that silencing of KLF2 significantly abolished OM stimulation-induced expression of Alp, Osx, and Ocn at the protein level; d Alizarin Red S staining assay revealed that silencing of KLF2 reduced osteogenic medium (OM)-induced mineralization of MC3T3-E1 cells; e ALP activity (*P < 0.01 vs. NS group; # P < 0.01 vs. NS + OM group)
KLF2 was then overexpressed in MC3T3-E1 cells by transduction with lentivirus encoding for KLF2. Successful overexpression of KLF2 is shown in Fig. 5a. Overexpression of KLF2 significantly enhanced OM stimulation-induced expression of Alp, Osx, and Ocn at both the mRNA level (Fig. 5b) and the protein level (Fig. 5c). Alizarin Red S staining results indicate that overexpression of KLF2 promoted OM-induced mineralization of MC3T3-E1 cells (Fig. 5d). MC3T3-E1 cells were transduced with lentivirus encoding for KLF2, followed by stimulation with OM for 14 days. ALP activity in MC3T3-E1 cells was then examined to evaluate osteoinductive activity (Fig. 5e). The results indicated that overexpression of KLF2 promoted an OM-induced increase in ALP activity. These findings implicate that KLF2 can stimulate osteoblast differentiation.
Overexpression of KLF2 promotes osteoblast differentiation. MC3T3-E1 cells were transduced with lentivirus encoding for KLF2. EV, empty virus; lenti-KLF2, lentivirus encoding for KLF2. a Western blot analysis revealed the successful overexpression of KLF2; b Real-time PCR analysis revealed that overexpression of KLF2 significantly promoted osteogenic medium stimulation-induced expression of Alp, Osx, and Ocn at the mRNA level; c Western blot analysis revealed that overexpression of KLF2 significantly promoted osteogenic medium stimulation-induced expression of Alp, Osx, and Ocn at the protein level; d Alizarin Red S staining assay revealed that overexpression of KLF2 promoted osteogenic medium (OM)-induced mineralization of MC3T3-E1 cells; e ALP activity (*P < 0.01 vs. EV group; #P < 0.01 vs. EV + OM group)
To further confirm the effects of KLF2 on osteoblastic differentiation, we overexpressed KLF2 in bone marrow mesenchymal stem cells. Western blot analysis revealed the successful expression of KLF2 in bone marrow mesenchymal stem cells (supplementary Fig. 2A). As expected, overexpression of KLF2 in bone marrow mesenchymal stem cells significantly increased the expression of Runx2 (supplementary Fig. 2B) as well as the expressions of Alp, Osx, and Ocn (supplementary Fig. 2C). Correspondingly, Alizarin Red S staining assay demonstrated that overexpression of KLF2 promoted OM-induced mineralization of bone marrow mesenchymal stem cells (supplementary Fig. 2D). These findings implicate that KLF2 can stimulate osteoblast differentiation.
We next investigated the impact of KLF2 on the expression of Runx2. Indeed, we found that silencing of KLF2 reduced the expression of Runx2 at both the mRNA level (Fig. 6a) and the protein level (Fig. 6b). Additionally, overexpression of KLF2 remarkably promoted the expression of Runx2 at both the mRNA level (Fig. 6c) and protein level (Fig. 6d).
KLF2 increased the expression of Runx2 with or without osteogenic medium (OM) treatment. a Real-time PCR analysis revealed that knockdown of KLF2 significantly reduced the expression of Runx2 at the mRNA level; b Western blot analysis revealed that knockdown of KLF2 significantly reduced the expression of Runx2 at the protein level; c Real-time PCR analysis revealed that overexpression of KLF2 significantly promoted the expression of Runx2 at the mRNA level; d Western blot analysis revealed that overexpression of KLF2 significantly promoted the expression of Runx2 at the protein level (*, #, $, P < 0.01)
Increasing evidence has shown that Runx2 cooperates with other regulatory proteins to regulate osteoblast differentiation. Therefore, immunoprecipitation experiments were performed to assess whether KLF2 could interact with Runx2. Flag-tagged KLF2 or Myc-tagged Runx2 were transfected into HEK-293T cells with constructs. Proteins were extracted and immunoprecipitated with Myc or KLF2 antibodies. The results in Fig. 7a indicate that exogenously overexpressed KLF2 and Runx2 could physically interact with each other in cells. Additionally, immunoprecipitation assay indicated that endogenous KLF2 could interact with Runx2 in pre-osteoblast MC3T3-E1 cells (Fig. 7b).
KLF2 physically interacted with Runx2. a HEK-293T cells were transfected with Flag-KLF2 and Myc-Runx2. Cell lysates were immunoprecipitated with Myc or Flag antibodies followed by western blot analysis with Flag, Runx2, KLF2, or Runx2 antibody; b Cell lysates from MC3T3-E1 cells were immunoprecipitated with KLF2 antibody followed by western blot analysis with Runx2 antibody
We then investigated the influence of KLF2 on Runx2 activity in MC3T3-E1 cells using Runx2 binding osteoblast-specific genes with luciferase reporter. The results indicate that overexpression of KLF2 alone had no significant influence on the luciferase activity of Alp (Fig. 8a), Osx (Fig. 8b), and Ocn (Fig. 8c). However, when cells were co-transfected with KLF2 and Runx2, the results indicate that overexpression of KLF2 promoted Runx2-induced luciferase activity of Alp (Fig. 8a), Osx (Fig. 8b), and Ocn (Fig. 8c) in MC3T3-E1 cells. These findings implicate that KLF2 is capable of regulating osteoblast-specific gene expression through interacting with Runx2.
KLF2 enhanced Runx2 transactivity in MC3T3-E1 cells. MC3T3-E1 cells were overexpressed with KLF2 or Runx2 and transfected with Alp luciferase vector, Osx luciferase vector, or Ocn luciferase vector. a Luciferase activity of Alp; b Luciferase activity of Osx; c Luciferase activity of Ocn (*P < 0.01 vs. vehicle group; #P < 0.01 vs. Runx2 overexpression group)
Discussion
It is well-recognized that KLF2 is an anti-inflammatory and anti-oxidant molecule [15]. Previous studies have identified KLF2 as a “molecular switch” regulating critical aspects of vascular function and disease [16]. Thus, KLF2 has been considered as a novel therapeutic target for inflammatory-related diseases. Conventional deficiency of KLF2 is lethal, as mouse embryos do not survive past E12.5 to E14.5 due to intraembryonic and intraamniotic hemorrhage [17]. However, we note that that study did not mention the underlying mechanism involved in skeletal anomaly induced by KLF2 deficiency. In the current study, we report for the first time to our knowledge that KLF2 is expressed in osteoblasts. Moreover, we found that KLF2 expression was significantly increased during the osteoblastic differentiation process in pre-osteoblast MC3T3-E1 cells and primary osteoblasts. Notably, our results display that KLF2 could promote the expression of the osteoblastic differentiation marker genes Alp, Osx, and Ocn and stimulate mineralization by enhancing the expression of and interacting with Runx2. In contrast, si-RNA transfection knockdown of KLF2 produced the opposite effects.
Runx2 is a master transcription factor controlling osteoblast differentiation, matrix production, and mineralization during bone formation [18]. Activation of Runx2 plays a critical role in regulating the expression of osteoblast-specific genes such as Alp, Osx, and Ocn [19]. It has been reported that overexpression of Runx2 in adipose tissue-derived mesenchymal stem cells accelerated osteoblast differentiation by promoting osteoblastic gene expression, increasing Alp activity, and stimulating mineralization [20]. Runx2 also plays a critical role in normal bone development. Decreased expression of Runx2 has been associated with abnormal bone development and the pathogenesis of osteoporosis [21]. Although KLF proteins represent an abundant protein superfamily in mammals, relatively few KLFs related to osteogenesis have been functionally characterized. Since KLF2 is involved in osteoblast function and Runx2 is a master regulator of osteogenesis, it is crucial to determine whether KLF2 acts upstream or downstream of Runx2. Our transient overexpression and knockdown data demonstrate that Runx2 was up-regulated by KLF2 overexpression and down-regulated by KLF2 knockdown in MC3T3-E1 cells. Meanwhile, we found that KLF2-overexpressing cells displayed enhanced osteogenic differentiation potential. In contrast, silencing of KLF2 impaired the differentiation capacity of MC3T3-E1 cells. Importantly, knockdown of Runx2 abolished KLF2-induced promotion of osteogenic differentiation, the expression of osteogenic differentiation marker genes, such as Alp, Osx, and Ocn, as well as osteogenic mineralization (data not shown). These results suggest that KLF2 is a factor operating upstream of Runx2, and Runx2 up-regulation by KLF2 is sufficient to prime for osteogenesis. Runx2 activity is precisely regulated by transcriptional factors through protein–DNA or protein–protein interactions [22]. For example, the nuclear protein Stat1 can inhibit nuclear localization of Runx2 by directly interacting with Runx2 [23]. Another transcriptional factor Nrf2 can inhibit Runx2-dependent stimulation of Ocn promoter activity by interacting with Runx2 [24]. Myeloid Elf-1 like factor (MEF) forms a complex with Runx2 to interfere with binding of Runx2 to the OC promoter at the OSE2 site. Interestingly, KLF2 could also interact with cofactors to regulate cellular activity. KLF2 has been reported to interact with NF-κB to counteract some of the pro-inflammatory actions of NF-κB [25]. In the present study, we found that KLF2 could enhance the transcriptional activity of Runx2 on Alp, Osx, and Ocn by physical interaction with Runx2. Because both Runx2 and KLF2 have numerous co-factors, we speculated that perhaps both of these transcription factors interact with other proteins within the cell. It is well-known that Alp activity is up-regulated in the early stage of osteoblast differentiation. Increased Alp activity has been shown to promote bone matrix mineralization [26]. Consistently, we found that overexpression of KLF2 promoted mineralization of MC3T3-E1 cells while silencing of KLF2 prevented mineralization of MC3T3-E1 cells.
KLF2 and other two KLF family members, KLF4 and KLF5, have been reported to play a key role in regulating the self renewal of embryonic stem cells (hESCs) and the maintenance of ESC pluripotency [27]. Hall and colleagues found that KLF2 was included in a list of ESC-enriched genes, and conferred resistance to BMP-induced differentiation [28]. Importantly, KLF2 overexpression in human bone marrow stromal cells (hBMSCs) significantly affected their biological activity, resulting in increased proliferation and pluripotency in an ex vivo culture system, implicating that KLF2 may exert positive effects on the stemness and self-renewal potential of hBMSCs [29]. Here, we demonstrated that KLF2 promoted osteoblast differentiation by increasing Runx2 expression and physically interacting with Runx2. Due to the essential roles of osteogenic differentiation in maintaining bone integrity, we speculated that KLF2 might be a novel therapeutic target for bone disease.
References
Boyle WJ, Simonet WS, Lacey DL. Osteoclast differentiation and activation. Nature. 2003;423:337–42.
Ikeda K, Takeshita S. Factors and mechanisms involved in the coupling from bone resorption to formation: how osteoclasts talk to osteoblasts. J Bone Metab. 2014;21:163–7.
Rachner TD, Khosla S, Hofbauer LC. Osteoporosis: now and the future. Lancet. 2011;377:1276–87.
Valenti MT, Dalle Carbonare L, Mottes M. Osteogenic differentiation in healthy and pathological conditions. Int J Mol Sci. 2016;18:E41.
Komori T. Roles of Runx2 in Skeletal Development. Adv Exp Med Biol. 2017;962:83–93.
Komori T, Yagi H, Nomura S, Yamaguchi A, Sasaki K, Deguchi K, et al. Targeted disruption of Cbfa1 results in a complete lack of bone formation owing to maturational arrest of osteoblasts. Cell. 1997;89:755e64.
Yang JW, Jeong BC, Park J, Koh JT. PHF20 positively regulates osteoblast diferentiation via increasing the expression and activation of Runx2 with enrichment of H3K4me3. Sci Rep. 2017;7:8060.
Novodvorsky P, Chico TJ. The role of the transcription factor KLF2 in vascular development and disease. Prog Mol Biol Transl Sci. 2014;124:155–88.
McConnell BB, Yang VW. Mammalian Kruppel-like factors in healthand diseases. Physiol Rev. 2010;90:1337–81.
SenBanerjee S, Lin Z, Atkins GB, Greif DM, Rao RM, Kumar A, et al. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–15.
Shi H, Sheng B, Zhang F, Wu C, Zhang R, Zhu J, et al. Kruppel-like factor 2 protects against ischemic stroke by regulating endothelial blood brain barrier function. Am J Physiol Heart Circ Physiol. 2013;304:H796–805.
Yuan Y, Tan H, Dai P. Krüppel-Like Factor 2 Regulates Degradation of Type II Collagen by Suppressing the Expression of Matrix Metalloproteinase (MMP)-13. Cell Physiol Biochem. 2017;42:2159–68.
Bakker A, Klein-Nulend J. Osteoblast isolation from murine calvariae and long bones. Methods Mol Med. 2003;80:19–28.
Sila-Asna M, Bunyaratvej A, Maeda S, Kitaguchi H, Bunyaratavej N. Osteoblast differentiation and bone formation gene expression in strontium-inducing bone marrow mesenchymal stem cell. Kobe J Med Sci. 2007;53:25–35.
Shi J, Zhou LR, Wang XS, Du JF, Jiang MM, Song Z, et al. KLF2 attenuates bleomycin-induced pulmonary fibrosis and inflammation with regulation of AP-1. Biochem Biophys Res Commun. 2017;S0006-291X:32092–2.
Atkins GB, Jain MK. Role of Kruppel-like transcription factors in endothelial biology. Circ Res. 2007;100:1686–95.
Wani MA, Means RT Jr, Lingrel JB. Loss of LKLF function results in embryonic lethality in mice. Transgenic Res. 1998;7:229–38.
Liu TM, Lee EH. Transcriptional regulatory cascades in Runx2-dependent bone development. Tissue Eng Part B Rev. 2013;19:254–63.
Jeong JH, Choi JY. Interrelationship of Runx2 and estrogen pathway in skeletal tissues. BMB Rep. 2011;44:613–8.
Zhang X, Yang M, Lin L, Chen P, Ma KT, Zhou CY, et al. Runx2 overexpression enhances osteoblastic differentiation and mineralization in adipose--derived stem cells in vitro and in vivo. Calcif Tissue Int. 2006;79:169–78.
Komori T. Signaling networks in RUNX2-dependent bone development. J Cell Biochem. 2011;112:750–5.
Schroeder TM, Jensen ED, Westendorf JJ. Runx2: a master organizer of gene transcription in developing and maturing osteoblasts. Birth Defects Res C Embryo Today. 2005;75:213–25.
Kim S, Koga T, Isobe M, Kern BE, Yokochi T, Chin YE, et al. Stat1 functions as a cytoplasmic attenuator of Runx2 in the transcriptional program of osteoblast differentiation. Genes Dev. 2003;17:1979–91.
Hinoi E, Fujimori S, Wang L, Hojo H, Uno K, Yoneda Y. Nrf2 negatively regulates osteoblast differentiation via interfering with Runx2-dependent transcriptional activation. J Biol Chem. 2006;281:18015–24.
SenBanerjee S, Lin Z, Atkins GB, et al. KLF2 Is a novel transcriptional regulator of endothelial proinflammatory activation. J Exp Med. 2004;199:1305–15.
Chin A, Yang Y, Chai L, Wong RW, Rabie AB. Effects of medicinal herb salvia miltiorrhiza on osteoblastic cells in vitro. J Orthop Res. 2011;29:1059–63.
Jiang J, Chan YS, Loh YH, Cai J, Tong GQ, Lim CA, et al. A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat Cell Biol. 2008;10:353–60.
Hall J, Guo G, Wray J, Eyres I, Nichols J, Grotewold L, et al. Oct4 and LIF/Stat3 additively induce Krüppel factors to sustain embryonic stem cell self-renewal. Cell Stem Cell. 2009;5:597–609.
Wang H, Zhou Y, Yu D, Zhu H. Klf2 contributes to the stemness and self-renewal of human bone marrow stromal cells. Cytotechnology. 2016 ;68:839–48.
Acknowledgements
This research was supported by the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), the Jiangsu Provinceʼs Key Medical center, Jiangsu Provincial Special Program of Medical Science (BL2012004), the Jiangsu Provincial Medical Youth Talent (QNRC2016751), the Natural Science Foundation of Jiangsu province (BK20180001), and the application fundamental research program of Suzhou City (SYSD2016072, SYS2018032).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Hou, Z., Wang, Z., Tao, Y. et al. KLF2 regulates osteoblast differentiation by targeting of Runx2. Lab Invest 99, 271–280 (2019). https://doi.org/10.1038/s41374-018-0149-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-018-0149-x
This article is cited by
-
Cyclin A1 (CCNA1) inhibits osteoporosis by suppressing transforming growth factor-beta (TGF-beta) pathway in osteoblasts
BMC Musculoskeletal Disorders (2024)
-
Targeting strategies for bone diseases: signaling pathways and clinical studies
Signal Transduction and Targeted Therapy (2023)
-
Neurodevelopmental disorders and cancer networks share pathways, but differ in mechanisms, signaling strength, and outcome
npj Genomic Medicine (2023)
-
Ferutinin induces osteoblast differentiation of DPSCs via induction of KLF2 and autophagy/mitophagy
Cell Death & Disease (2022)
-
scATAC-Seq reveals heterogeneity associated with spermatogonial differentiation in cultured male germline stem cells
Scientific Reports (2022)