Abstract
Downregulation of deleted in liver cancer 1 (DLC1) is associated with poor prognosis of various cancers, but its functional mechanisms in hepatocellular carcinoma (HCC) remains unclear. In the present study, we investigated the roles of DLC1 in tumor progression and autophagy of HCC. We found that DLC1 was frequently downregulated in HCC tissues. Underexpression of DLC1 correlated with AFP level, vascular invasion, poor differentiation, and poor prognosis. In vitro assays revealed that DLC1 not only suppressed the proliferation, migration, and invasion of HCC cells, but also inhibited autophagy of HCC cells. Mechanistic investigation revealed that DLC1 decreased TCF4 expression and the interaction between β-catenin and TCF4, then inactivated Wnt/β-catenin signaling. Additionally, DLC1 suppressed the ROCK1 activity and the dissociation of the Beclin1-Bcl2 complex, thereby inhibiting autophagy of HCC cells. In conclusion, our findings imply that loss of DLC1 contributes to the progression and oncogenic autophagy of HCC.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is the fifth most common malignancies and the third leading cause of cancer-related deaths around the world [1]. Despite the significant improvement of surgical and adjuvant treatment approaches, the 5-year survival rate of HCC remains unsatisfactory due to the the high rate of intrahepatic and extrahepatic metastases [2]. Therefore, enhancing our understanding of the molecular mechanisms in HCC is urgently needed for promoting the development of effective therapy and improving the prognosis of patients with HCC.
Human deleted in liver cancer 1 (DLC1) is a bona fide tumor suppressor gene and downregulated via genetic and epigenetic mechanisms in variety of cancers, including lung caner, breast cancer, gastric cancer, and liver cancer [3,4,5,6]. DLC1 is a multidomain protein that contains sterile alpha motif (SAM), Rho GTPase-activating protein (RhoGAP), and steroidogenic acute regulatory protein-related lipid transfer (START) domains. DLC1 mainly acts as a GAP for RhoA, RhoB, RhoC, and Cdc42 [7]. The RhoGAP activity of DLC1 negatively regulates these Rho proteins by stimulating their intrinsic GTP hydrolytic activity, thus converts them from the active GTP-bound state to the inactive GDP-bound state. The Rho signaling cascade controls many biological processes, such as cell proliferation and the cytoskeleton organization [8, 9]. Rho GTPases and DLC1 are dysregulated in several cancers and have been found to play opposite roles in tumorigenesis and tumor metastasis [10, 11].
Wnt/β-catenin signaling is involved in the regulation of a number of processes, including embryonic phase cell multiplication, stem cell proliferation, organogenesis, and cellular migration [12]. Previous studies identified that Wnt/β-catenin signaling pathway was deregulated in patients with HCC, resulting in increased cell proliferation, migration, and invasion [13, 14]. However, a complete knowledge of the regulatory relationship between DLC1 and Wnt/β-catenin signaling pathway in HCC is not well understood yet.
Autophagy is an evolutionarily conserved process in which intracellular membrane structures sequester proteins and intracellular components to degrade and recycle these materials to maintain cellular homeostasis and regain energy [15]. Emerging evidences imply the significance of autophagy in tumor initiation and development. Autophagy contributes to cell survival via reproduction of nutrients and restoration of metabolic stress, but its excess also results in the type 2 cell death (autophagic cell death) [16]. Thus, the role of autophagy is controversial in tumor pathogenesis. It is generally believed that autophagy acts as a tumor suppressor at the early stage of liver carcinogenesis [17], while autophagy promotes HCC progression at advanced stages of tumor development [18]. However, the association between DLC1 and autophagy modulation in HCC remains unclear.
In this study, we systematically investigated the function of DLC1 in the tumorigenesis and progression of HCC. We found that DLC1 expression was downregulated in the majority of HCC tissues. Downregulation of DLC1 was significantly associated with alpha-fetoprotein (AFP) level, vascular invasion, low-level differentiation, and poor prognosis in HCC patients. Moreover, functional assays showed that DLC1 significantly suppressed tumor growth, migration, invasion, and autophagy of HCC. Mechanistically, DLC1 repressed the Wnt/β-catenin signaling and the dissociation of the Beclin1-Bcl2 complex. These results provide a new understanding of the underlying mechanism by which DLC1 inhibits HCC.
Materials and methods
Cell culture and tissue samples
SK-Hep-1, Huh-7, HepG2, SMMC-7721, and Hep3B cells were obtained from the cell bank of the Institute of Biochemistry and Cell Biology of the Chinese Academy of Sciences (Shanghai, China). Authentication of these cell lines was performed using the GenePrint10 System (Promega Biotech Co.) and via comparisons to the STR database. MHCC-97h cells were obtained from Zhongshan Hospital of Fudan University. Cells were cultured in DMEM (Hyclone) containing 10% fetal bovine serum (PAN Biotech), 100 U/ml penicillin, and 100 mg/ml streptomycin at 37 °C.
A total of 94 pairs of HCC tissue and corresponding non-tumor tissue samples were collected from HCC patients who initially underwent hepatectomy without any preoperative treatment at the Zhongshan Hospital of Xiamen University from 2011 to 2015. The use of human material was approved by the ethics committee of the Zhongshan Hospital of Xiamen University, and written informed consent was obtained from these subjects.
DLC1 knockdown and overexpression
Short hairpin RNA (shRNA) target sequences were referenced from a previous study [19] and provided as follows: shDLC1: CCTTGACTGGAATATGTAA. Scramble shRNA was taken as a negative control, and the sequence of hairpin is: CCTAAGGTTAAGTCGCCCTCGCTCGAGCGAGGGCGACTTAACCTTAGG. Sramble and DLC1 shRNAs were cloned into pLKO.1 vector. Wild-type DLC1 or mutant DLC1 (R178E) was cloned into pLV vector. These lentiviral expressing vectors were co-transfected with lentiviral packaging vectors (pMD2.G and psPAX2) into 293T cells (ATCC) by using TurboFect Transfection Reagent (Thermo Scientific) according to the the manufacturers’ instructions. Lentiviral particles were collected after 48 h. Cells were transfected with lentiviral constructs as described above for 24 h. Stable cells were selected with puromycin for 5 days. The overexpression and knockdown effect was determined by western blot.
Cell proliferation and colony formation assays
Cell proliferation was determined by cell counting kit-8 (CCK-8) assay. A total of 2.0 × 103 cells per well were seeded in 96-well culture plates. At the indicated time points, CCK-8 (Dojindo) was added to each well and incubated at 37 °C for 90 min. The absorbance values (OD 450 nm) were measured using a spectrophotometer (Bio-Rad). For colony formation assay, 2 × 103 cells per well were seeded in the six-well culture plates. After 14 days’ culture, cells were fixed with 4% paraformaldehyde and stained with crystal violet.
Migration and invasion assays
For cell migration and invasion assays, 24-well Transwell chambers with 8 µm pore size polycarbonate membrane were used (Corning, NY, USA). Cells were planted on the top side of the membrane precoated with Matrigel (BD Bioscience, NJ, USA; without Matrigel for cell migration assay) and incubated for 24 h. Cells inside the upper chamber were obliterated with cotton swabs, while cells on the lower membrane surface were fixed and then stained with 0.5% crystal violet solution. Five fields were counted randomly in each well.
Animal experiments
A xenograft mouse model was developed using male BALB/C nude mice. Stable cells were trypsinized and suspended in 100 μl serum-free DMEM. Cells were injected subcutaneously into the right flank of the nude mice. Tumors were detected after ~14 days, and tumor size was measured every 3 days. Tumor-bearing mice were killed 38 days after the injection. All animal experiments were approved by the Animal Care and Use Committee of the Xiamen University.
RNA isolation and real-time PCR
Total RNA was isolated using TRIzol (Invitrogen) according to the manufacturers’ instructions. Complementary DNA (cDNA) was synthesized from total RNA using one-step gDNA Removal and cDNA Synthesis Kit (Transgen, Beijing, China). Real-time PCR was performed on a Lightcycle™ Real-Time PCR System (Roche) using FastStart Universal SYBR Green Master (Rox) (Roche). Data were normalized to GAPDH or to control samples. Comparative quantification was determined using the 2−ΔΔCt method. The primers for the detected genes were shown as follow: DLC1-F: GCGTACCTGTGTCGCTTTAT; DLC1-R: CTCCTCTGTGCAAACCTTTCT; TCF4-F: CACTTTCCCTAGCTCCTTCTTC; TCF4-R: TGCCCAACATTCCTGCATAG; GAPDH-F: CTTTGGTATCGTGGAAGGACTC; GAPDH-R: AGTAGAGGCAGGGATGATGT.
Immunohistochemistry
Four-micrometer sections were pretreated before immunohistochemistry (IHC) staining. After deparaffinization of slides in xylene and rehydration through a graded ethanol series, an antigen retrieval step was performed. Endogenous peroxidase was quenched by incubation of the sections with 0.3% hydrogen peroxide for 15 min at room temperature. Non-specific binding was blocked by incubation with nonimmune serum for 15 min at room temperature. The sections were then incubated with anti-DLC1 antibody (1:200, sc-32931, Santa Cruz) at 4 °C overnight. After wash, the sections were incubated with biotinylated secondary antibody (KIT-5010, Maixin Biotechnology, Fuzhou, China), and then visualized using diaminobenzidine (Maixin Biotechnology, Fuzhou, China).
Immunofluorescence
Samples were fixed with 4% paraformaldehyde for 10 min. Subsequently, the cells were permeabilized with 0.1% Triton X-100 for 10 min at room temperature, washed with PBS, and blocked with PBS containing 1% (w/v) bovine serum albumin for 30 min at room temperature. Samples were incubated with the LC3B (#3868, Cell Signaling Technology) antibodies overnight at 4 °C, washed with PBS, and incubated with corresponding secondary IgG (Alexa Fluor® 488, A-11034, ThermoFisher) for 1 h at room temperature. After rinsing in PBS, the samples were counterstained with diamidino phenylindole (DAPI) (Sigma). Representative images were photographed using a confocal microscope.
Luciferase activity
β-catenin/TCF firefly luciferase reporter construct (pTOPflash) and pRL-TK reporter plasmid encoding Renilla luciferase was purchased from Promega (Madison, WI, USA). Cells were seeded onto 12-well plates at 6 × 104 cells per well and were transiently transfected with 0.4 µg of pTOPflash along with 0.04 µg of pRL-TK using Turbofect (Thermo) following the manufacturer’s protocol. The cells were collected 24 h post transfection. After lysed and centrifuged, and the supernatant was used for the measurement of luciferase activities using the Dual-Luciferase Assay System (Promega). The firefly luciferase activity was normalized to Renilla luciferase activity and was expressed as the relative luciferase activity.
Co-immunoprecipitation
Cells were lysed with immunoprecipitation (IP) lysis buffer (Beyotime). Total protein was incubated with 50 μl of Protein G Sepharose (GE healthcare) for 1 h at 4 °C on a rocking platform to reduce non-specific binding. After removing the beads, the supernatant was incubated with 5 μg anti-TCF4 (#2565, Cell Signaling Technology), anti-β-catenin (#8480, Cell Signaling Technology), anti-Bcl2 (12789-1-AP, Proteintech), anti-Beclin1 (#3495, Cell Signaling Technology) antibodies, or negative control anti-rabbit IgG (#2729, Cell Signaling Technology) overnight at 4 ℃. A total of 50 μl of protein G sepharose was added to each immunoprecipitation mixture, and the incubation was continued for 1 h at 4 °C on a rocking platform. The immunoprecipitates were collected and washed three times with the cold IP lysis buffer. After the loading buffer was added, the sepharose was boiled and subjected to western blot assay. The clean-blot IP detection reagent (horse radish peroxidase) secondary antibodies (21232, Thermo) were employed to avoid the denatured heavy and light chains from antibodies used in co-IP assays.
Western blot
Cells or tissues were lysed with RIPA buffer (Beyotime) containing protease inhibitors cocktail (Selleck). Protein samples were loaded onto and separated using SDS-PAGE, then transferred onto polyvinylidene fluoride membranes (Milliore). The membranes were blocked for 1 h at room temperature and incubated with the anti-LC3B (#3868, Cell Signaling Technology), anti-ACTB (#3700, Cell Signaling Technology), anti-TCF4 (#2565, Cell Signaling Technology), anti-β-catenin (#8480, Cell Signaling Technology), anti-Bcl2 (12789-1-AP, Proteintech), or anti-Beclin1 (#3495, Cell Signaling Technology) antibodies overnight at 4 °C. After washing, the blots were incubated with goat anti-rabbit (111-035-003, Jackson) or anti-mouse (115-035-003, Jackson) horse radish peroxidase-conjugated secondary antibodies and visualized using the Immobilon™ horse radish peroxidase substrate peroxid solution (Millipore).
Statistical analysis
All statistical analyses in our experiment were performed using the SPSS version 19.0 software system (IBM, Armonk, NY, USA). Data are shown as mean ± SEM. The differences between groups were analyzed by the Student’s t-test or χ2-test. The Kaplan–Meier method was performed for patients’ overall survival analysis. All experiments were performed in triplicate. P < 0.05 was considered statistically significant difference.
Results
Underexpression of DLC1 is associated with clinicopathological features and poor prognosis of patients with HCC
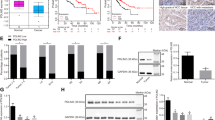
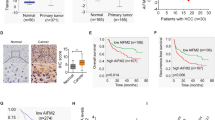
To compare the expression pattern of DLC1 in HCC tumor tissues and adjacent nonneoplastic tissues, we detected the protein levels of DLC1 in 94 pairs of HCC tumor and matched non-tumor specimens by IHC assays. We found that DLC1 was significantly decreased in 64.9% (61/94) HCC tumor tissues compared to matched non-tumor tissues (Fig. 1a). We also compared the DLC1 expression in 10 pairs of focal nodular hyperplasia and normal liver tissues, but no difference was observed (Supplemental Fig. 1). Western blotting analysis revealed that DLC1 expression can be detected in the majority of non-tumoral liver specimens, but a significant portion of patients showed decreased DLC1 level in HCC tumor tissues (21 of 35 cases) (Fig. 1b). Moreover, analysis of DLC1 messenger RNA (mRNA) expression from 29 patients with HCC by RT-PCR showed that average DLC1 mRNA levels was much lower in tumor tissues than non-tumor liver tissues (P < 0.0001, Fig. 1c). These data suggested that downregulation of DLC1 may implicated in HCC carcinogenesis and progression.
DLC1 is significantly downregulated in HCC tissues. a IHC analysis of DLC1 expression in 94 pairs of HCC tumor tissues (T) and matched non-tumoral liver tissues (N). b The DLC1 protein levels in 35 pairs of HCC tumor tissues (T) and matched non-tumoral liver tissues (N) was determined by western blot. The representative image was shown. c The expression of DLC1 mRNA levels in 28 pairs of HCC tumor tissues (T) and matched non-tumoral liver tissues (N) was detected by RT-PCR. d Kaplan–Meier’s analyses were performed according to DLC1 protein expression on HCC patients. T ≥ N, the expression of DLC1 was not downregulated in HCC tumor tissues compared to non-tumoral liver tissues. T < N, the expression of DLC1 was downregulated in HCC tumor tissues compared to non-tumoral liver tissues
To determine the clinical significance of DLC1 expression, we analyzed the correlation between DLC1 expression and clinicopathological features of patients with HCC through IHC assays (Fig. 1a). As shown in Table 1, downregulation of DLC1 expression in HCC tissues was significantly correlated with higher serum AFP level (P = 0.024), vascular invasion (P = 0.002), and low-level differentiation (P = 0.011). However, no associations were identified between DLC1 expression and other clinical features, such as age, gender, hepatitis B virus infection, and tumor size. Moreover, downregulation of DLC1 in HCC was found to predict shorter overall survival (P = 0.032, Fig. 1d) of patients. Taken together, these findings highlighted the clinical relevance of DLC1 in HCC pathogenesis.
DLC1 negatively regulates HCC growth in vitro and in vivo
We first determined the expression of DLC1 using a panel of HCC cell lines. The level of DLC1 was low in MHCC-97h cells, whereas DLC1 is highly expressed in SK-Hep-1 cells (Fig. 2a). We then analyzed the functional role of DLC1 in HCC initiation and progression by knockdown and overexpression approaches. We first used lentiviral constructs to stably silence DLC1 in SK-Hep-1 cells and overexpressed DLC1 in MHCC-97h cells (Fig. 2b). CCK-8 assays showed that DLC1-silencing cells grew much faster than control cells (Fig. 2c) and overexpression of DLC1 significantly inhibited cell proliferation (Fig. 2d). In addition, we found that DLC1-knockdown SK-Hep-1 cells enhanced colony-forming ability (Fig. 2e), whereas overexpression of DLC1 remarkably reduced MHCC-97h cell clonogenicity (Fig. 2f). To determine whether Rho inhibition mediates the suppressive role of DLC1 in cell proliferation, we stably overexpressed the R718E DLC1 mutant (point substitution in the RhoGAP domain to abolish its catalytic activity on GTPases) [20] and compared its function with overexpression of wild-type DLC1 in MHCC-97h cells. Concordantly, the RhoGAP mutation completely abolished the suppressive function of DLC1 in cell proliferation and clonogenicity (Fig. 2d, f).
Effect of DLC1 on the growth of HCC cells in vitro and in vivo. a Expression of DLC1 in different HCC cell lines was determined by western blot. ACTB was used as a loading control. b Construct stable MHCC-97h cells with DLC1 overexpression and SK-Hep-1 cells with DLC1 knockdown. c Proliferation of SK-Hep-1 cells with or without DLC1 knockdown was assessed by CCK-8 assays. d Proliferation of MHCC-97h cells expressing wild-type DLC1 or mutant DLC1 (R718E) was assessed by CCK-8 assays. e Clone formation assay of SK-Hep-1 cells with or without DLC1 knockdown. f Clone formation assay of MHCC-97h cells expressing wild-type DLC1 or mutant DLC1 (R718E). g DLC1 stable clones were inoculated into nude mice, and tumor size was measured. All quantitative data shown represent the means ± SEM of at least three independent experiments. *P < 0.05
The inhibitory function of DLC1 in HCC tumorigenicity was further confirmed in vivo by a subcutaneous xenograft model. Control, wild-type DLC1 and R718E mutant overexpressing MHCC-97h cells were subcutaneously injected into nude mice, and tumor growth was measured. Consistent with our in vitro data, wild-type DLC1 efficiently suppressed tumor formation compared to control and R718E cells (Fig. 2g). Collectively, DLC1 exerts suppressive function in HCC growth in vitro and in vivo through its catalytic activity of RhoGAP domain.
DLC1 represses cell migration and invasion in HCC
In addition to cell proliferation, our clinicopathological analysis showed that downregulation of DLC1 in HCC was significantly associated with the presence of vascular invasion, which is a well-established indicator of HCC metastasis. Therefore, we examined the effect of DLC1 on migratory and invasive ability through transwell assays. The results showed that SK-Hep-1 cells with DLC1 knockdown migrated and invaded faster than control cells (Fig. 3a). In addition, overexpression of wild-type DLC1, but not of R718E DLC1, suppressed migration and invasion of MHCC-97h cells (Fig. 3b).
The effect of DLC1 on cell migration and invasion. a The migration and invasive ability after knockdown of DLC1 in SK-Hep-1 cells was assessed using transwell assays. b The migration and invasive ability of MHCC-97h cells expressing DLC1 or R718E was assessed by transwell assays. All data shown represent the means ± SEM of at least three independent experiments. *P < 0.05
DLC1 suppresses Wnt/β-catenin signaling
Considering the significant role of Wnt/β-catenin signaling in regulating cells proliferation and migration, we hypothesized Wnt/β-catenin signaling pathway involved in the biological effects of DLC1 in the progression of HCC. To assure whether DLC1 affected WNT/β-catenin pathway, TOP/FOP luciferase reporter system of common WNT pathway activation was introduced in our study. Our results showed that WNT signaling was activated by DLC1 knockdown (Fig. 4a), while WNT signaling was suppressed by DLC1 overexpression (Fig. 4b).
DLC1 suppresses Wnt/β-catenin signaling. a Luciferase activity of TOPflash reporter was evaluated in SK-Hep-1 cells with or without DLC1 knockdown. b Luciferase activity of TOPflash reporter was evaluated in MHCC-97h cells with or without DLC1 overexpression. c Stable cell clones were fractionated into cytoplasmic and nuclear portions followed by immunoblotting using antibodies against β-catenin, lamin B1, and α-tubulin. d The mRNA and protein levels of TCF4 in SK-Hep-1 cells with or without DLC1 knockdown were detected by RT-PCR and western blot, respectively. e The mRNA and protein levels of TCF4 in MHCC-97h cells with or without DLC1 overexpression were detected by RT-PCR and western blot, respectively. f DLC1 was overexpressed in MHCC-97h cells, and after immunoprecipitating endogenous TCF4, bound β-catenin was subjected to western blotting. All quantitative data shown represent the means ± SEM of at least three independent experiments. *P < 0.05
Previous study demonstrated that DLC1 associated with α-catenin, E-cadherin, and β-catenin, constitutive adherens junctions proteins [21]. Binding of DLC1 to α-catenin led to their accumulation at the plasma membrane, suggesting that DLC1 may influence WNT signaling through inhibiting β-catenin nuclear accumulation. Unfortunately, using cytoplasmic and nuclear protein fractions from HCC cells, we observed that neither DLC1 knockdown or overexpression have an effect on the nuclear translocation of β-catenin (Fig. 4c). We then analyzed the mRNA expression profiles mediated by DLC1 from a recent study [19]. Among these mRNA candidates, T-cell factor 4 (TCF4) draw our attention for its combination with β-catenin to activate Wnt signaling. To validate this result, we examined the expression level of TCF4 in control and DLC1-knockdown cells through RT-PCR and western blot. We observed that both mRNA and protein level of TCF4 was increased by DLC1 downregulation (Fig. 4d), whereas overexpression of DLC1 significantly suppressed TCF4 expression (Fig. 4e). We speculated DLC1 might inhibit the formation of β-catenin-TCF4 complex. The co-IP experiment demonstrated that the interaction of endogenous β-catenin and TCF4 was significantly decreased by DLC1 overexpression (Fig. 4f). These results demonstrate that DLC1 suppresses the activation of Wnt signaling through downregulation of TCF4 expression.
DLC1 inhibits autophagy in HCC
Finally, we investigated whether DLC1 have an effect on autophagy in human HCC cells. Knockdown of DLC1 caused increased autophagic flux as evidenced by conversion from LC3-I to LC3-II and degradation of p62 in SK-Hep-1 cells (Fig. 5a), while overexpression of DLC1 in MHCC-97h cells reversed it (Fig. 5b). We further investigated whether DLC1 has a role in autophagy by examining the morphological features of endogenous LC3 (seen as puncta). As shown in Fig. 5c, DLC1 silencing significantly increased LC3 puncta accumulation in both the cytoplasm and nucleus. In contrast, the amount of LC3 dots per cell was significantly decreased in DLC1-overexpressing cells as compared with the control group.
DLC1 inhibits autophagy in HCC cells. a The protein levels of LC3 and p62 in SK-Hep-1 cells with or with our DLC1 knockdown were analyzed by western blot. Representative western blot and densitometric analysis normalized to ACTB demonstrating the effect of DLC1 silencing on LC3-II levels. b The protein levels of LC3 and p62 in MHCC-97h cells with or withour DLC1 overexpression were analyzed by western blot. Representative western blot and densitometric analysis normalized to ACTB demonstrating the effect of DLC1 upregulation on LC3-II levels. c Representative immunofluorescent images showing redistribution of autophagic marker LC3 in DLC1-knockdown SK-Hep-1 cells were taken on a confocal microscope. d Representative immunofluorescent images showing redistribution of autophagic marker LC3 in DLC1-overexpressing MHCC-97h cells were taken on a confocal microscope. All quantitative data shown represent the means ± SEM of at least three independent experiments. *P < 0.05
DLC1 attenuates the interaction between Beclin1 and Bcl2 via inactivating ROCK1
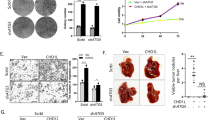
Beclin1 has a central role during the autophagy process, which was initially identified as interacting partner of the anti-apoptotic protein, Bcl-2 [22, 23]. Beclin1 binds Bcl-2 and inhibits autophagosome formation, while Beclin1 is released from Bcl-2 and then proceed to exert its function in autophagy [24]. A recent study reported that activation of Rho-associated kinase 1 (ROCK1) decreased the Beclin1-Bcl2 association, thus inducing autophagy [25]. As shown in Fig. 6a, knockdown of DLC1 was able to phosphorylate MYPT1, a known substrate of ROCK1, as compared with control cells, while overexpression of DLC1 decreased the phosphorylation level of MYPT1. In addition, enzyme-linked immunosorbent assay using MYPT1 as a substrate showed that ROCK1 activity is significantly elevated in DLC1-silencing SK-Hep-1 cells and reduced in DLC1-overexpressing MHCC-97h cells compared to their control cells, respectively (Fig. 6b, c). These results demonstrated that DLC1 suppressed the activity of ROCK1 in HCC cells.
DLC1 attenuates the interaction between Beclin1 and Bcl2 to repress autophagy in HCC cells. a Endogenous phosphorylation of MYPT1 was determined by western blotting. b SK-Hep-1 cells with or without DLC1 knockdown were lysed, extracts normalized, and ROCK1 kinase activity was measured by performing enzyme-linked immunosorbent assay. c MHCC-97h cells with or without DLC1 overexpression were lysed, extracts normalized, and ROCK1 kinase activity was measured by performing enzyme-linked immunosorbent assay. d The effect of DLC1 overexpression on the interaction between Bcl2 and Beclin1 was detected by co-IP assay. e The effect of DLC1 knockdown on the interaction between Bcl2 and Beclin1 was detected by co-IP assay. All quantitative data shown represent the means ± SEM of at least three independent experiments. *P < 0.05
As DLC1 negatively regulates ROCK1 activity, we hypothesized that DLC1 may inhibit the Beclin1-Bcl2 interaction through suppression of ROCK1 activation in HCC. To test this hypothesis, we performed co-IP experiments and observed that Beclin1 binding to Bcl2 was promoted after DLC1 overexpression (Fig. 6d). In contrast, DLC1 knockdown repressed the association between Beclin1 and Bcl2 (Fig. 6e). Taken together, our findings indicate that DLC1 suppress autophagy through inhibiting the ROCK1 activity and the disruption of Beclin1-Bcl2 complex.
Discussion
Downregulation of DLC1 was found in various human cancers due to gene deletion or epigenetic silence, such as lung cancer, breast cancer, and liver cancer [6, 26]. In our previous study, we also demonstrated that DLC1 transcription was silenced by insulin-like growth factor 1 (IGF1) pathway-mediated DNA methylation [5]. Aberrant DLC1 expression is associated with poor prognosis of patients with cancer [27, 28]. However, to date, the clinicopathological and prognostic significance of DLC1 expression in HCC remains unknown. Consistent with previous studies, we observed that both mRNA and protein levels in HCC tissues were significantly decreased compared to adjacent non-tumor liver tissues. Correlation analysis showed that DLC1 expression in HCC tissues was associated with AFP level, vascular invasion, and poor differentiation. Moreover, downregulation of DLC1 predicted poor prognosis of HCC patients. These findings indicate that DLC1 may be a potential indicator for prognostic assessment of HCC.
Rho GTPases constitute a distinct family of small GTPases within the Ras superfamily. Rho GTPases are key regulators of the actin and microtubule cytoskeleton, thereby controlling different steps of cell migration, cell cycle progression, and differentiation [29]. Aberrant Rho signaling in cancer is caused by altered gene expression or function of their regulators. For example, many RhoGEFs such as Dbl and Ect2 were identified to be upregulated or mutated, leading to altered signaling and malignant transformation [30, 31]. In addition, suppression of GAP activity alone is sufficient to cause the hyperactivation of Rho [32]. DLC1 plays an outstanding role in cancer, since inactivation is the most common alteration reported for Rho regulators. In our present study, loss- and gain-of-function assays showed that knockdown of DLC1 promoted cell proliferation, migration, and invasion, while overexpression of DLC1 reversed these phenotypes. The RhoGAP mutation (R718E) completely abolished the suppressive function of DLC1 in cell proliferation, migration, and invasion, suggested that the RhoGAP activity was essential for the tumor suppressive function of DLC1.
Wnt signaling is one of the most important molecular pathways in cancer growth and metastasis. A previous study demonstrated that DLC1 inhibited the growth, invasion, and Wnt pathway through suppression of β-catenin in colon cancer [33]. However, our results showed that overexpression or knockdown of DLC1 did not influence the the nuclear translocation and the expression of β-catenin. Our results showed that DLC1 overexpression significantly inhibited the β-catenin-TCF4 interaction, suggesting that DLC1 regulated Wnt/β-catenin signaling in a tissue-specific manner. TCF4 is the most important transcription factor in the TCF/LEF family. TCF4 binds to DNA and regulates the transcription of a variety of target genes. Interaction between TCF4 and β-catenin is responsible for activation of transcription of many tumor-related proteins, including c-myc, cyclin D1, and ZEB1, and play essential roles in tumorigenesis and development of many cancers [34,35,36]. In the present study, the findings of RT-PCR and western blot assays verified the downregulation of TCF4 in DLC1-overexpressing cells. Conversely, knockdown of DLC1 increased TCF4 expression. The mechanisms by which DLC1 regulated TCF4 transcription need further investigation.
Rho-associated kinase (ROCK) is the fundamental downstream effector of the small GTPase Rho, which have a role in various cellular processes, including cell adhesion, migration, invasion, transformation, mitosis, DNA damage, and apoptosis [37,38,39]. Overexpression of ROCK significantly contributes to metastasis by enhancing tumor cell invasion and motility [40]. Moreover, activated ROCK1 promotes autophagy by binding and phosphrylating Beclin1 and dissociation of the Beclin1-Bcl2 complex [25]. It has been reported that ectopic expression of DLC1 abrogated ROCK-mediated cytoskeletal reorganization including formation of stress fibers and focal adhesions in HCC cells, and suppressed metastasis through negatively regulating Rho/ROCK pathway in prostate cancer [41, 42]. However, the role of DLC1-ROCK1 pathway in autophagy of HCC cells is still unclear. For the first time, we illuminate the role of DLC1 deregulation in autophagy in HCC. Our data demonstrated that knockdown of DLC1 activated ROCK1 and promoted the dissociation of the Beclin1-Bcl2 complex, which induced autophagy in HCC cells. Conversely, overexpression of DLC1 showed the opposite effects. Overall, our present data further the understanding of DLC1 in HCC pathogenesis and progression.
References
Torre LA, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108.
Budhu A, Forgues M, Ye QH, et al. Prediction of venous metastases, recurrence, and prognosis in hepatocellular carcinoma based on a unique immune response signature of the liver microenvironment. Cancer Cell. 2006;10:99–111.
Park H, Cho SY, Kim H, et al. Genomic alterations in BCL2L1 and DLC1 contribute to drug sensitivity in gastric cancer. Proc Natl Acad Sci USA. 2015;112:12492–7.
Low JS, Tao Q, Ng KM, et al. A novel isoform of the 8p22 tumor suppressor gene DLC1 suppresses tumor growth and is frequently silenced in multiple common tumors. Oncogene. 2011;30:1923–35.
Fang QL, Yin YR, Xie CR, et al. Mechanistic and biological significance of DNA methyltransferase 1 upregulated by growth factors in human hepatocellular carcinoma. Int J Oncol. 2015;46:782–90.
Wang D, Qian X, Rajaram M, Durkin ME, Lowy DR. DLC1 is the principal biologically-relevant down-regulated DLC family member in several cancers. Oncotarget. 2016;7:45144–57.
Goodison S, Yuan J, Sloan D, et al. The RhoGAP protein DLC-1 functions as a metastasis suppressor in breast cancer cells. Cancer Res. 2005;65:6042–53.
Wang T, Kang W, Du L, Ge S. Rho-kinase inhibitor Y-27632 facilitates the proliferation, migration and pluripotency of human periodontal ligament stem cells. J Cell Mol Med. 2017;21:3100–12.
Jeong KJ, Park SY, Cho KH, et al. The Rho/ROCK pathway for lysophosphatidic acid-induced proteolytic enzyme expression and ovarian cancer cell invasion. Oncogene. 2012;31:4279–89.
Cardama GA, Gonzalez N, Maggio J, Menna PL, Gomez DE. Rho GTPases as therapeutic targets in cancer (review). Int J Oncol. 2017;51:1025–34.
Liao YC, Lo SH. Deleted in liver cancer-1 (DLC-1): a tumor suppressor not just for liver. Int J Biochem Cell Biol. 2008;40:843–7.
Bejsovec A. Flying at the head of the pack: Wnt biology in Drosophila. Oncogene. 2006;25:7442–9.
Peng YY, He YH, Chen C, et al. NLRC5 regulates cell proliferation, migration and invasion in hepatocellular carcinoma by targeting the Wnt/beta-catenin signaling pathway. Cancer Lett. 2016;376:10–21.
Pez F, Lopez A, Kim M, et al. Wnt signaling and hepatocarcinogenesis: molecular targets for the development of innovative anticancer drugs. J Hepatol. 2013;59:1107–17.
Kondo Y, Kanzawa T, Sawaya R, Kondo S. The role of autophagy in cancer development and response to therapy. Nat Rev Cancer. 2005;5:726–34.
Galluzzi L, Vicencio JM, Kepp O, et al. To die or not to die: that is the autophagic question. Curr Mol Med. 2008;8:78–91.
Takamura A, Komatsu M, Hara T, et al. Autophagy-deficient mice develop multiple liver tumors. Genes Dev. 2011;25:795–800.
Luo T, Fu J, Xu A, et al. PSMD10/gankyrin induces autophagy to promote tumor progression through cytoplasmic interaction with ATG7 and nuclear transactivation of ATG7 expression. Autophagy. 2016;12:1355–71.
Wang Y, Lei R, Zhuang X, et al. DLC1-dependent parathyroid hormone-like hormone inhibition suppresses breast cancer bone metastasis. J Clin Invest. 2014;124:1646–59.
Healy KD, Hodgson L, Kim TY, et al. DLC-1 suppresses non-small cell lung cancer growth and invasion by RhoGAP-dependent and independent mechanisms. Mol Carcinog. 2008;47:326–37.
Tripathi V, Popescu NC, Zimonjic DB. DLC1 interaction with alpha-catenin stabilizes adherens junctions and enhances DLC1 antioncogenic activity. Mol Cell Biol. 2012;32:2145–59.
Liang XH, Kleeman LK, Jiang HH, et al. Protection against fatal Sindbis virus encephalitis by beclin, a novel Bcl-2-interacting protein. J Virol. 1998;72:8586–96.
Maiuri MC, Le Toumelin G, Criollo A, et al. Functional and physical interaction between Bcl-X(L) and a BH3-like domain in Beclin-1. EMBO J. 2007;26:2527–39.
Pattingre S, Tassa A, Qu X, et al. Bcl-2 antiapoptotic proteins inhibit Beclin 1-dependent autophagy. Cell. 2005;122:927–39.
Gurkar AU, Chu K, Raj L, et al. Identification of ROCK1 kinase as a critical regulator of Beclin1-mediated autophagy during metabolic stress. Nat Commun. 2013;4:2189.
Liu Y, Zhou J, Hu Y, Wang J, Yuan C. Curcumin inhibits growth of human breast cancer cells through demethylation of DLC1 promoter. Mol Cell Biochem. 2017;425:47–58.
Castro M, Grau L, Puerta P, et al. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med. 2010;8:86.
Qian X, Durkin ME, Wang D, et al. Inactivation of the Dlc1 gene cooperates with downregulation of p15INK4b and p16Ink4a, leading to neoplastic transformation and poor prognosis in human cancer. Cancer Res. 2012;72:5900–11.
Braun AC, Olayioye MA. Rho regulation: DLC proteins in space and time. Cell Signal. 2015;27:1643–51.
Lebowitz PF, Prendergast GC. Functional interaction between RhoB and the transcription factor DB1. Cell Adhes Commun. 1998;6:277–87.
Chen J, Xia H, Zhang X, et al. ECT2 regulates the Rho/ERK signalling axis to promote early recurrence in human hepatocellular carcinoma. J Hepatol. 2015;62:1287–95.
Vincent S, Settleman J. Inhibition of RhoGAP activity is sufficient for the induction of Rho-mediated actin reorganization. Eur J Cell Biol. 1999;78:539–48.
Wang C, Wang J, Liu H, Fu Z. Tumor suppressor DLC-1 induces apoptosis and inhibits the growth and invasion of colon cancer cells through the Wnt/beta-catenin signaling pathway. Oncol Rep. 2014;31:2270–8.
He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–12.
Saegusa M, Hashimura M, Kuwata T, Hamano M, Okayasu I. Beta-catenin simultaneously induces activation of the p53-p21WAF1 pathway and overexpression of cyclin D1 during squamous differentiation of endometrial carcinoma cells. Am J Pathol. 2004;164:1739–49.
Sanchez-Tillo E, de Barrios O, Siles L, et al. Beta-catenin/TCF4 complex induces the epithelial-to-mesenchymal transition (EMT)-activator ZEB1 to regulate tumor invasiveness. Proc Natl Acad Sci USA. 2011;108:19204–9.
Riento K, Ridley AJ. Rocks: multifunctional kinases in cell behaviour. Nat Rev Mol Cell Biol. 2003;4:446–56.
Ongusaha PP, Qi HH, Raj L, et al. Identification of ROCK1 as an upstream activator of the JIP-3 to JNK signaling axis in response to UVB damage. Sci Signal. 2008;1:ra14.
Schofield AV, Bernard O. Rho-associated coiled-coil kinase (ROCK) signaling and disease. Crit Rev Biochem Mol Biol. 2013;48:301–16.
Liu S. The ROCK signaling and breast cancer metastasis. Mol Biol Rep. 2011;38:1363–6.
Huang W, Liu J, Feng X, et al. DLC-1 induces mitochondrial apoptosis and epithelial mesenchymal transition arrest in nasopharyngeal carcinoma by targeting EGFR/Akt/NF-kappaB pathway. Med Oncol. 2015;32:115.
Tripathi V, Popescu NC, Zimonjic DB. DLC1 induces expression of E-cadherin in prostate cancer cells through Rho pathway and suppresses invasion. Oncogene. 2014;33:724–33.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (no. 81672418, no. 81702351, no. 81702348, and no. 81372618,), the Major State Basic Research Development Program of China (grant no. 2017YFA0205201), the Fujian Youth Key Personnel Research Project (no. 2013-ZQN-ZD-34 and 2018-ZQN-84), the Science and Technology Project of Xiamen (no. 3502Z20164022), the Fujian Provincial Health and Family Planning Youth Research Project (no. 2018-2-64) and the Natural Science Foundation of Fujian (no. 2018J01389 and 2014D022).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wu, HT., Xie, CR., Lv, J. et al. The tumor suppressor DLC1 inhibits cancer progression and oncogenic autophagy in hepatocellular carcinoma. Lab Invest 98, 1014–1024 (2018). https://doi.org/10.1038/s41374-018-0062-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41374-018-0062-3
This article is cited by
-
LINC00114 stimulates growth and glycolysis of esophageal cancer cells by recruiting EZH2 to enhance H3K27me3 of DLC1
Clinical Epigenetics (2022)
-
MiR-200a-3p promotes gastric cancer progression by targeting DLC-1
Journal of Molecular Histology (2022)
-
Frequent Downregulation and Promoter Hypermethylation of DLC1: Relationship with Clinical Outcome in Gallbladder Cancer
Journal of Gastrointestinal Cancer (2022)
-
Authentication of differential gene expression in oral squamous cell carcinoma using machine learning applications
BMC Oral Health (2021)
-
CircZKSCAN1 Suppresses Hepatocellular Carcinoma Tumorigenesis by Regulating miR-873-5p/Downregulation of Deleted in Liver Cancer 1
Digestive Diseases and Sciences (2021)