Abstract
Intervertebral disc degeneration is a major cause of low back pain. The nucleus pulposus (NP) is an important intervertebral disc component. Recent studies have shown that carbonic anhydrase 12 (CA12) is a novel NP marker. However, the mechanism by which CA12 is regulated and its physiological function are unclear. In our study, CA12, hypoxia-inducible factor 1α (HIF-1α) and HIF-2α expression levels were examined in 81 human degenerated NP samples using real-time RT-PCR, immunohistochemistry and western blot. Rat NP cells were cultured in a hypoxic environment, and hypoxia-induced CA12 expression was examined. Rat NP cells were treated with HIF-1α siRNA or the prolyl hydroxylase (PHD) inhibitor dimethyloxalylglycine (DMOG) to evaluate the role of PHD/HIF-1 in regulating CA12 expression. Rat NP cells were treated with CA12 siRNA to determine the function of CA12. A rat ex vivo model was established to confirm that PHD, HIF-1, and CA12 have important roles in disc degeneration. We found that CA12 was significantly downregulated in degenerated human NP samples at the mRNA and protein levels. CA12 expression sharply increased by ~30-fold in response to hypoxia. The expression of HIF-1α, but not HIF-2α, also decreased in degenerated human NP samples and was positively correlated with CA12 expression. HIF-1α knockdown under hypoxia reduced the CA12 mRNA and protein expression levels. DMOG treatment increased HIF-1α and CA12 expression. CA12 knockdown significantly inhibited anabolic protein expression, whereas catabolic enzymes remained unchanged. The ex vivo experiments supported our in vitro studies of the role of PHD/HIF-1/CA12. In conclusion, CA12 is downregulated in degenerated NPs, and its expression may be regulated by the PHD/HIF-1 axis. Decreased CA12 expression may lead to decreased extracellular matrix synthesis, which contributes to degenerative disc disease progression.
Similar content being viewed by others
Main
Low back pain (LBP) is one of the most common clinical complaints. Approximately 80% of people experience LBP during their lifespan,1 and this condition causes severe pain, disability, and heavy economic burden. Among the various causes of LBP, intervertebral disc degeneration is thought to be one of the most central causes.2, 3 The intervertebral disc consists of the central proteoglycan-rich nucleus pulposus (NP) and the surrounding fibrocartilaginous annulus fibrosis (AF) with good restoration of strength, ROM, and function. The highly absorptive proteoglycans in the NP absorb stress and maintain the structural and functional integrity of the spine.4, 5 An imbalance between NP extracellular matrix (ECM) anabolism and catabolism leads to degenerative disc disease.6 NP cell aging or changes to the microenvironment can lead to decreased normal ECM protein synthesis (mainly type II collagen and aggrecan) and increased synthesis of ECM proteins related to fibrosis, such as type I collagen.6 Inflammatory cytokines, such as interleukin-1β and tumor necrosis factor-α, stimulate the expression of ECM catabolic enzymes, such as matrix metalloproteinases and a disintegrin and metalloproteinase with thrombospondin motifs (ADAMTSs).7, 8 Loss of ECM results in reduced disc height and altered mechanical characteristics of the spine, leading to progressive degenerative disc disease.
Carbonic anhydrase 12 (CA12) is a transmembrane protein of the carbonic anhydrase family, which includes zinc metalloenzymes that catalyze reversible CO2 hydration into bicarbonate and hydrogen ions.9 Hypoxia-induced CA12 upregulation promotes tumor survival and growth by maintaining intracellular pH.10, 11, 12 Microarray and real-time RT-PCR results have confirmed the significant differential expression of CA12 in NP compared with articular cartilage and AF.13, 14 Several research groups have begun to use CA12 as an NP marker.15, 16, 17 Thus, we hypothesize that CA12 has a role in NP physiology. Although Power et al14 found that CA inhibition disrupts pH and leads to lactate accumulation in NP cells, the exact function of CA12 in NP remains unknown. CA12 regulation in the NP is also unknown. Because CA12 expression is induced by hypoxia18 in tumor cells and because the intervertebral disc is a nonvascular and relatively hypoxic organ, we hypothesize that hypoxia regulates CA12 expression in the NP. Furthermore, we suggest that CA12 has an important role in the NP degeneration process and that its expression is regulated by the prolyl hydroxylase (PHD)/hypoxia-inducible factor (HIF) axis. In this study, we aimed to confirm the change in CA12 expression in degenerated NPs, determine the mechanism of CA12 regulation in the NP in vitro and ex vivo, and elucidate the function of CA12 in the NP in vitro and ex vivo.
Materials and methods
Human Tissue Collection and Grading
Informed consent was obtained from each patient. Our study was approved by the Institutional Review Board of Sir Run Run Shaw Hospital. NP samples were obtained from patients with degenerative disc disease who were undergoing discectomy and fusion (degenerated group) or patients with vertebral burst fractures who underwent discectomy and fusion but did not have a history of LBP (control group). The degenerated group was further subdivided into three subgroups (Grade III, Grade IV, and Grade V) according to the Pfirrmann grading system.19 The patients’ clinical characteristics are summarized in Table 1. All samples were collected in our department between January 2012 and December 2013. After collection, samples were immediately dissected under a microscope and either frozen in liquid nitrogen or fixed in 4% buffered paraformaldehyde for 24 h.
Rat NP Cell Culture and Treatment
The NP was separated from the lumbar discs of 8-week-old male Sprague–Dawley rats using a dissecting microscope and finely diced into small pieces no >1 mm3 in size. The tissues were treated with 0.2% type II collagenase (Sigma-Aldrich, St. Louis, MO, USA) for 1 h at 37 °C. The supernatant was placed into Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 20% fetal bovine serum (Gibco, Gaithersburg, MD, USA) in a humidified atmosphere containing 5% CO2 at 37 °C. For the hypoxia treatment, cells were cultured in a hypoxic incubator with a mixture of 1% O2, 5% CO2, and 94% N2 for 6–72 h. CA12 or hypoxia-inducible factor 1α (HIF-1α) expression was downregulated by treating NP cells with CA12 or HIF-1α siRNA, respectively (Genepharm, Shanghai, China). In brief, cells were grown to 50% confluence and transfected with chemically synthesized target siRNA or negative control siRNA using Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. To determine the role of PHD, we treated cells with 0.5, 1, or 2 mM of the competitive PHD inhibitor dimethyloxalylglycine (DMOG) (Sigma-Aldrich) for 24 h.
Isolation and Culture of Disc/Endplate Specimens
Thoracic and lumbar intervertebral discs, including the adjacent vertebral endplates, were separated from 8-week-old male Sprague–Dawley rats. Specimens were washed in Hank’s solution (Solarbia, Beijing, China) with 55 mM Na-citrate (Sigma-Aldrich). Discs were washed with agitation overnight in DMEM supplemented with 5% fetal calf serum and 20 mM Na-citrate in an incubator (37 °C, 5% CO2, 100% humidity). Specimens were treated with or without 1 mM DMOG and 100 μM of the CA inhibitor acetazolamide (ACTZ) (Sigma-Aldrich) for up to 2 weeks in 48-well plates with DMEM supplemented with 10% fetal calf serum and 25 μg/ml l-ascorbate.
Real-time RT-PCR Analysis
Total RNA was extracted from NP tissues or the treated NP cells, cultured as described above. NP tissues that had been frozen in liquid nitrogen were ground into a fine powder using a mortar and pestle, and total RNA was extracted using the RNeasy Mini kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s protocol. Complementary DNA (cDNA) was synthesized from 500 ng RNA from each sample, 2 μl 5 × PrimeScript RT Master Mix (Takara Bio, Otsu, Japan) and RNase-free dH2O in a final volume of 10 μl. Real-time RT-PCR was performed using a 96-well plate ABI Prism 7500 (Applied Biosystems, Foster City, CA, USA) with SsoFast EvaGreen Supermix (Bio-Rad, Hercules, CA, USA). The total volume (20 μl) of each PCR reaction contained 10 μl SsoFast EvaGreen Supermix, 7 μl RNase-free dH2O, 2 μl cDNA, and 10 μM of each forward and reverse primer (Table 2). The quantity of each target gene was normalized to β-actin expression.
Western Blot
Cells or ground NP tissues were incubated in RIPA buffer (Cell Signaling Technology, Boston, MA, USA) supplemented with 100 mM phenylmethanesulfonyl fluoride (PMSF) on ice, followed by centrifugation at 12 000 rpm for 15 min to isolate the supernatant. Proteins were resolved by 10% SDS-PAGE and transferred to PVDF membranes (Bio-Rad) by electroblotting. The membranes were blocked with 5% non-fat dry milk in TBST at room temperature for 1 h and then incubated with anti–CA12 (1:1000; Cell Signaling Technology), anti-HIF-1α (1:1000; Santa Cruz Biotechnology, Santa Cruz, CA, USA) or anti-β-actin (1:2000; Santa Cruz Biotechnology) polyclonal immunoglobulin G antibodies overnight at 4 °C. Protein bands were developed using a horseradish peroxidase-conjugated goat-anti-rabbit immunoglobulin G (Abcam, Cambridge, MA, USA), followed by detection with ECL reagent (Millipore, Billerica, MA, USA). Protein bands were visualized using the LAS-4000 Science Imaging System (Fujifilm, Tokyo, Japan).
Histological Studies
Tissues were fixed in 4% buffered paraformaldehyde for 24 h. Rat disc samples were decalcified in 10% EDTA for 1 month. Tissues were then dehydrated in graded ethanol solutions and embedded in paraffin. For each specimen, three serial sections (4-μm thick) were cut on a microtome. To observe morphology and matrix degeneration, sections were stained with hematoxylin-eosin (H&E) or safranin O (SO)-fast green. To examine CA12 immunoreactivity, an immunohistochemical study was performed using a histostain SABC kit (CWBIO, Beijing, China) according to the manufacturer’s instructions. Samples without treatment of CA12 antibody was included as negative control. Specimens were examined and photographed using a high-quality microscope.
Statistical Analysis
Statistical analyses were performed using SPSS for Windows software, version 16.0 (SPSS, Chicago, IL, USA). All data are presented as the mean±s.e.m. A Mann–Whitney U-test was used to compare the mean values of two groups. A Spearman rho test was used to analyze the correlation between transcriptional levels of different genes. P-values<0.05 were considered significant.
Results
Evaluation of 81 Human NP Degeneration Samples
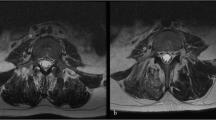
We performed MRI analysis, H&E staining, SO staining, and real-time RT-PCR and found significant degeneration in the degenerated group compared with the control group (Figures 1a and k). Compared with the control group MRI (Figures 1a and b), the degenerated group MRI (Figures 1c and d) showed decreased signal intensity of NP in T2WI, a narrower disc space and a disappearing margin between the NP and AF. As shown by H&E staining (Figures 1e and f), the cell numbers were significantly decreased, and the cell shapes changed from small and round (Figure 1e) to spindle-like (Figure 1f). There was also evidence of more hypertrophic cells in the degenerated group. Furthermore, SO staining (Figures 1g and h) demonstrated significantly decreased proteoglycan content in the degenerated group (Figure 1h) compared with the control group (Figure 1g). Real-time RT-PCR results showed decreased expression of type II collagen (Figure 1i), SOX9 (Figure 1j), and aggrecan (Figure 1k).
Evaluation of human NP samples by magnetic resonance image (MRI), histological study and real-time RT-PCR. (a, b) The T1WI (a) and T2WI (b) of a 41-year-old male patient with L1 vertebral fracture. The NP of L1/L2 was collected in our study (white arrow). (c, d) The T1WI (c) and T2WI (d) of a 61-year-old male patient with degenerative disc disease. The NP of L4/L5 was collected in our study (white arrow). (e–h) H&E staining of the control group (e mag. × 100) and degenerated group (f mag. × 100); safranin O staining of the control group (g mag. × 100) and degenerated group (h mag. × 100). Scale bar: 100 μm. i–k: Real-time RT-PCR results of type II collagen (i), SOX9 (j), and aggrecan (k) of the degenerated group and control group. **P<0.01.
CA12 and HIF-1α Expression was Downregulated in Degenerated NP Tissues
We examined CA12 transcription in the 81 NP samples and found that it was significantly downregulated in the degenerated groups (Figure 2a). We further divided the degenerated group into three subgroups according to the Pfirrmann grading system and found that CA12 expression progressively decreased from the control group to Grade V (Figure 2b). Immunohistochemical analysis (Figure 2c) showed that CA12 localized to the plasma membrane, and the number of CA12-positive cells decreased in the degenerated group. As observed by western blot, the protein levels (Figure 2d) were in accordance with the mRNA levels.
CA12 and HIF-1α expression decreased in the degenerated group. (a) Real-time RT-PCR detection of CA12 mRNA in the degenerated group and control group. (b) Real-time RT-PCR detection of CA12 mRNA in the degenerate group after subdivision by the Pfirrmann grading system. (c) Immunohistochemical detection of CA12 protein in human NP samples from both groups. Scale bar: 100 μm. (d) CA12 western blot analysis in both groups (C1, C2, and C3 from the control group; D1, D2, and D3 from the degenerated group). (e, f) Real-time RT-PCR detection of HIF-1α (e) and HIF-2α (f) expression in degenerated and control groups. *P<0.05; **P<0.01.
CA12 Expression was Regulated by PHD/HIF-1α in Hypoxic Conditions
To examine the relationship between CA12 and relative hypoxic conditions in NP cells, we measured CA12 expression under hypoxic conditions by real-time RT-PCR. As shown in Figure 3a, CA12 mRNA was significantly upregulated at 12, 24, 48, and 72 h under hypoxic conditions. The mRNA level was upregulated by ~30-fold by 72 h. Because the CA12 promoter contains two hypoxia-responsive elements (HRE, (A/G)CGTG), we hypothesized that hypoxia-induced factors regulate CA12 by directly binding to its promoter. To determine which HIF has a role in human NPs, we examined HIF-1α and HIF-2α expression levels in the 81 human NP samples. As shown in Figures 2e and f, HIF-1α mRNA levels significantly decreased, whereas HIF-2α levels remained unchanged. The HIF-1α mRNA level, but not the HIF-2α level, positively correlated with the CA12 mRNA level (Spearman coefficient 0.25; P<0.05). To determine whether HIF-1α regulates CA12 expression, we suppressed HIF-1α expression under hypoxic conditions using HIF-1α siRNA. Western blot (Figure 3b) and real-time RT-PCR (Figure 3c) confirmed that knockdown was efficient. Notably, CA12 expression was significantly decreased in cells that were transduced with HIF-1α siRNA in a dose-dependent manner (Figure 3d). In contrast, HIF-1α knockdown did not affect CA12 expression under normoxic conditions (Figure 3e). In addition, treatment with the PHD inhibitor DMOG significantly induced CA12 expression under normoxic conditions in a dose-dependent manner (Figure 3f).
CA12 expression was induced by hypoxia, and mediated by PHD/HIF-1α. (a) Real-time RT-PCR results of CA12 mRNA in rat NP cells treated under hypoxia for 0, 6, 12, 24, 48, or 72 h. (b) Western blot analysis of NP cells treated with negative control siRNA or 100 nM HIF-1α siRNA under hypoxic conditions. (c, d) Real-time RT-PCR detection of HIF-1α (c) and CA12 (d) in rat NP cells treated with 50, 100 or 200 nM HIF-1α siRNA or negative control siRNA under hypoxic conditions. (e) Real-time RT-PCR detection of CA12 in rat NP cells treated with negative control siRNA or 100 nM HIF-1α siRNA under normoxic conditions. (f) Real-time RT-PCR detection of CA12 in rat NP cells treated with 0, 0.5, or 2 mM DMOG. *P<0.05; **P<0.01.
CA12 Downregulation Altered ECM Synthesis
To identify the function of CA12 in intervertebral disc degeneration, we suppressed its expression using siRNA in rat NP cells under hypoxic conditions. As shown in Figures 4a and b, we confirmed CA12 downregulation at both the protein and mRNA levels. Upon CA12 knockdown, type II collagen and SOX9 levels significantly decreased (Figures 4c and d). Aggrecan levels also decreased, although without statistical significance (Figure 4f). Interestingly, type I collagen was upregulated after CA12 knockdown (Figure 4e). The expression of catabolic enzymes (eg, MMP3, ADAMTS-4, and ADAMTS-5) (Figures 4g and i) did not change.
The role of CA12 in NP anabolism and catabolism. (a) Western blot analysis of CA12 in rat NP cells treated with CA12 siRNA under hypoxic condition. (b) Real-time RT-PCR of CA12 in rat NP cells treated with CA12 siRNA under hypoxic conditions. (c–f) Real-time RT-PCR of type II collagen (c), SOX9 (d), type I collagen (e), and aggrecan (f) in rat NP cells treated with CA12 siRNA under hypoxic conditions. (g–i) Real-time RT-PCR of MMP3 (g), ADAMTS-4 (h), and ADAMTS-5 (i) in rat NP cells treated with CA12 siRNA under hypoxic conditions. *P<0.05; **P<0.01.
The Role of PHD/HIF-1α/CA12 in an ex vivo Rat Disc Model
To further explore the potential protective effects of PHD/HIF-1α/CA12 in degenerative disc disease, we used a whole disc ex vivo model. H&E staining and SO staining showed that the ECM was significantly degraded in the control group, as described by Haschtmann et al,20 and DMOG treatment prevented this degradation. When discs were treated with the CA inhibitor ACTZ, DMOG’s protective effect was dampened (Figures 5a and b). An analysis of the mRNA levels further confirmed these results. After a 2-week treatment with DMOG, aggrecan, type II collagen, and SOX9 expression significantly increased, whereas type I collagen expression decreased (Figures 5c and f). DMOG treatment also induced CA12 and HIF-1α expression (Figures 5g and i). However, when discs were co-treated with ACTZ and DMOG, type II collagen, type I collagen, aggrecan, and SOX9 expression were partially restored to control levels (Figures 5c and f).
The role of PHD/HIF-1α/CA12 in an ex vivo rat NP model. (a) H&E staining of sections of discs treated with 1 mM DMOG or 1 mM DMOG plus 100 μM ACTZ under normoxia. Scale bar: 100 μm. (b) SO staining of sections of discs treated with 1 mM DMOG or 1 mM DMOG plus 100 μM ACTZ under normoxia. Scale bar: 100 μm. (c–h) Real-time RT-PCR analysis of aggrecan (c), type II collagen (d), type I collagen (e), SOX9 (f), CA12 (g), and HIF-1α (h) in discs treated with 1 mM DMOG or 1 mM DMOG plus 100 μM ACTZ under normoxia. (i) Western blot analysis of CA12 and HIF-1α expression in discs treated with 1 mM DMOG or 1 mM DMOG plus 100 μM ACTZ under normoxia. *P<0.05; **P<0.01.
Discussion
CA12 is enriched in human NP tissue compared with other tissues, including AF and cartilage.13, 14 Our research further exhibited that CA12 was downregulated in the degenerated NP, suggesting that CA12 has a role in degenerative disc disease and may be a marker for disc degeneration. The trend of decreased CA12 expression among subgroups divided by the Pfirrmann grading system also suggested that CA12 levels in the NP could be used as a biomarker of degenerative disc disease severity and progression. These results raise two questions: what is the physiological function of CA12 in NP cells, and what is the primary mechanism that controls CA12 expression in NP cells?
Although limited vessels infiltrate the outer AF layers and cartilage endplate, they do not enter the NP.21 NP cells are in a hypoxic microenvironment under physiological conditions.22, 23 CA12 expression is induced by hypoxia in VHL-defective renal cell carcinoma,24 suggesting that it may be similarly regulated in NP cells. Our results showed that CA12 mRNA expression increased by ~30-fold after 72 h in hypoxic conditions.
HIF mediates oxygen homeostasis by regulating numerous enzymes in response to hypoxia.25, 26 HIFs are heterodimers that are composed of an oxygen tension-regulated α-subunit and a constitutively expressed β-subunit. The α-subunit translocates to the nucleus and forms a heterodimer with the β-subunit.27 The heterodimer then binds to a specific site in the promoter, termed the HRE, and regulates several genes that are important for cell survival under hypoxia, such as erythropoiesis, vascular endothelial growth factor, and glucose transporters.25, 28 However, several proline residues in the α-subunit are hydrolyzed by PHDs under normoxia. The α-subunit then becomes poly-ubiquitinated and degraded by the proteasome.27 It has been found that HIF-1 and HIF-2 are expressed in NP tissue.29, 30, 31 We found that the human CA12 promoter contains 2 HREs (5′-C/GCGTG-3′), and HIF-1 has been reported to mediate CA12 upregulation in tumor cells.18 These data suggest that decreased CA12 may also be regulated by HIF in the degenerated NP.
To determine which HIF functions in disc degeneration, we examined HIF-1α and HIF-2α expression in 81 NP tissues. Our results suggested that HIF-1α is significantly downregulated and that its expression positively correlated with CA12 expression. We further validated these findings by HIF-1α knockdown. Transfection with different doses of HIF-1α led to dose-dependent CA12 downregulation. However, HIF-1α did not modulate CA12 expression under normoxia, demonstrating the importance of PHDs in HIF-1α-induced CA12 expression. Therefore, we inhibited PHDs using the pan-PHD inhibitor DMOG under normoxia for 24 h and found that HIF-1α and CA12 expression were dose-dependently induced.
We next investigated CA12 function in NP cells. CAs are enzymes that catalyze the reversible hydration of cell-generated carbon dioxide into protons and bicarbonate ions.11, 32 Thus, CA12 can contribute to cellular alkalinization and intracellular pH homeostasis. Chiche et al18 and Doyen et al33 previously found that CA12 suppression decreases tumor cell survival by increasing intracellular acidosis. Power et al14 found that the CA12 inhibitor 4-aminobenzene sulfonamide decreases lactate levels in hypoxic NP cells, suggesting that CA12 can counteract cellular acidosis. We confirmed the function of CA12 in NP catabolism and anabolism by knocking down CA12 expression. Anabolic genes, such as type II collagen, SOX9 and aggrecan, were downregulated without affecting catabolic genes. Type I collagen, which is related to NP fibrosis and degeneration, was upregulated. Our data highlight the protective roles of CA12 during the process of disc degeneration.
In our ex vivo studies, we found that DMOG treatment increased anabolism, and this effect could be partially blocked by the CA inhibitor ACTZ. This finding was in accordance with our in vitro experiments. Because ACTZ is a carbonic anhydrase inhibitor, our results suggest that CA12 promotes anabolism during disc degeneration by regulating NP pH homeostasis.
Although our study highlights important findings, further studies are required. First, we were unable to examine direct interaction between HIF-1 and the CA12 promoter, although we showed that HIF-1 modulated CA12 expression. This can be determined in the future through chromatin immunoprecipitation studies. Second, considering the influence of trauma on NP, a control group of normal human disc samples would be more suitable than samples from fracture patients. We are currently collecting normal post-mortem NP samples and this new control group will be used in future studies.
In conclusion, we revealed that PHD/HIF-1/CA12 is a novel pathway that may affect disc degeneration by modulating ECM anabolism. Therefore, genetic or pharmacological modulation of PHD/HIF-1/CA12 activity may be a novel therapeutic approach to degenerative disc disease.
References
Ehrlich GE . Low back pain. Bull World Health Organ 2003;81:671–676.
Samartzis D, Karppinen J, Mok F, Fong DY, Luk KD, Cheung KM . A population-based study of juvenile disc degeneration and its association with overweight and obesity, low back pain, and diminished functional status. J Bone Joint Surg Am 2011;93:662–670.
Battie MC, Videman T, Levalahti E, Gill K, Kaprio J . Heritability of low back pain and the role of disc degeneration. Pain 2007;131:272–280.
Setton LA, Chen J . Mechanobiology of the intervertebral disc and relevance to disc degeneration. J Bone Joint Surg Am 2006;88 (Suppl 2):52–57.
Feng H, Danfelter M, Stromqvist B, Heinegard D . Extracellular matrix in disc degeneration. J Bone Joint Surg Am 2006;88 (Suppl 2):25–29.
Kepler CK, Ponnappan RK, Tannoury CA, Risbud MV, Anderson DG . The molecular basis of intervertebral disc degeneration. Spine J 2013;13:318–330.
Wang J, Markova D, Anderson DG, Zheng Z, Shapiro IM, Risbud MV . TNF-alpha and IL-1beta promote a disintegrin-like and metalloprotease with thrombospondin type I motif-5-mediated aggrecan degradation through syndecan-4 in intervertebral disc. J Biol Chem 2011;286:39738–39749.
Le Maitre CL, Hoyland JA, Freemont AJ . Catabolic cytokine expression in degenerate and herniated human intervertebral discs: IL-1beta and TNFalpha expression profile. Arthritis Res Ther 2007;9:R77.
Gilmour KM . Perspectives on carbonic anhydrase. Comp Biochem Physiol A Mol Integr Physiol 2010;157:193–197.
Ivanov SV, Kuzmin I, Wei MH et al. Down-regulation of transmembrane carbonic anhydrases in renal cell carcinoma cell lines by wild-type von Hippel-Lindau transgenes. Proc Natl Acad Sci USA 1998;95:12596–12601.
Ivanov S, Liao SY, Ivanova A et al. Expression of hypoxia-inducible cell-surface transmembrane carbonic anhydrases in human cancer. Am J Pathol 2001;158:905–919.
Chiche J, Ricci JE, Pouyssegur J . Tumor hypoxia and metabolism — towards novel anticancer approaches. Ann Endocrinol (Paris) 2013;74:111–114.
Minogue BM, Richardson SM, Zeef LA, Freemont AJ, Hoyland JA . Characterization of the human nucleus pulposus cell phenotype and evaluation of novel marker gene expression to define adult stem cell differentiation. Arthritis Rheum 2010;62:3695–3705.
Power KA, Grad S, Rutges JP et al. Identification of cell surface-specific markers to target human nucleus pulposus cells: expression of carbonic anhydrase XII varies with age and degeneration. Arthritis Rheum 2011;63:3876–3886.
van den Akker GG, Surtel DA, Cremers A et al. Novel immortal human cell lines reveal subpopulations in the nucleus pulposus. Arthritis Res Ther 2014;16:R135.
Hu X, Zhou Y, Zheng X et al. Differentiation of menstrual blood-derived stem cells toward nucleus pulposus-like cells in a coculture system with nucleus pulposus cells. Spine (Phila Pa 1976) 2014;39:754–760.
Sun Z, Liu ZH, Zhao XH et al. Impact of direct cell co-cultures on human adipose-derived stromal cells and nucleus pulposus cells. J Orthop Res 2013;31:1804–1813.
Chiche J, Ilc K, Laferriere J et al. Hypoxia-inducible carbonic anhydrase IX and XII promote tumor cell growth by counteracting acidosis through the regulation of the intracellular pH. Cancer Res 2009;69:358–368.
Pfirrmann CW, Metzdorf A, Zanetti M, Hodler J, Boos N . Magnetic resonance classification of lumbar intervertebral disc degeneration. Spine (Phila Pa 1976) 2001;26:1873–1878.
Haschtmann D, Stoyanov JV, Ettinger L, Nolte LP, Ferguson SJ . Establishment of a novel intervertebral disc/endplate culture model: analysis of an ex vivo in vitro whole-organ rabbit culture system. Spine (Phila Pa 1976) 2006;31:2918–2925.
Rudert M, Tillmann B . Lymph and blood supply of the human intervertebral disc. Cadaver study of correlations to discitis. Acta Orthop Scand 1993;64:37–40.
Urban JP, Smith S, Fairbank JC . Nutrition of the intervertebral disc. Spine (Phila Pa 1976) 2004;29:2700–2709.
Urban JP . The role of the physicochemical environment in determining disc cell behaviour. Biochem Soc Trans 2002;30 (Pt 6):858–864.
Wykoff CC, Beasley NJ, Watson PH et al. Hypoxia-inducible expression of tumor-associated carbonic anhydrases. Cancer Res 2000;60:7075–7083.
Semenza GL . HIF-1: mediator of physiological and pathophysiological responses to hypoxia. J Appl Physiol (1985) 2000;88:1474–1480.
Carmeliet P, Dor Y, Herbert JM et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature 1998;394:485–490.
Kurihara T, Westenskow PD, Friedlander M . Hypoxia-inducible factor (HIF)/vascular endothelial growth factor (VEGF) signaling in the retina. Adv Exp Med Biol 2014;801:275–281.
Semenza GL, Wang GL . A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol Cell Biol 1992;12:5447–5454.
Fujita N, Chiba K, Shapiro IM, Risbud MV . HIF-1alpha and HIF-2alpha degradation is differentially regulated in nucleus pulposus cells of the intervertebral disc. J Bone Miner Res 2012;27:401–412.
Richardson SM, Knowles R, Tyler J, Mobasheri A, Hoyland JA . Expression of glucose transporters GLUT-1, GLUT-3, GLUT-9 and HIF-1alpha in normal and degenerate human intervertebral disc. Histochem Cell Biol 2008;129:503–511.
Ha KY, Koh IJ, Kirpalani PA et al. The expression of hypoxia inducible factor-1alpha and apoptosis in herniated discs. Spine (Phila Pa 1976) 2006;31:1309–1313.
Swietach P, Vaughan-Jones RD, Harris AL . Regulation of tumor pH and the role of carbonic anhydrase 9. Cancer Metastasis Rev 2007;26:299–310.
Doyen J, Parks SK, Marcie S, Pouyssegur J, Chiche J . Knock-down of hypoxia-induced carbonic anhydrases IX and XII radiosensitizes tumor cells by increasing intracellular acidosis. Front Oncol 2012;2:199.
Acknowledgements
This study is supported by the Chinese National Natural Science Foundation (81271971, 81301585, 81301587, 81472064), Zhejiang Provincial Natural Science Foundation of China (LZ15H060002).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
The nucleus pulposus (NP) is important in intervertebral disc degeneration, which leads to low back pain. This paper shows that carbonic anhydrase 12 (CA12), a novel NP marker, is down-regulated in degenerated NPs by the prolyl hydroxylase/HIF-1 axis, and may lead to decreased extracellular matrix synthesis.
Rights and permissions
About this article
Cite this article
Chen, S., Fang, XQ., Wang, Q. et al. PHD/HIF-1 upregulates CA12 to protect against degenerative disc disease: a human sample, in vitro and ex vivo study. Lab Invest 96, 561–569 (2016). https://doi.org/10.1038/labinvest.2016.32
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2016.32
This article is cited by
-
The role of HIF proteins in maintaining the metabolic health of the intervertebral disc
Nature Reviews Rheumatology (2021)
-
The RCAN1.4-calcineurin/NFAT signaling pathway is essential for hypoxic adaption of intervertebral discs
Experimental & Molecular Medicine (2020)
-
Expression of Carbonic Anhydrase III, a Nucleus Pulposus Phenotypic Marker, is Hypoxia-responsive and Confers Protection from Oxidative Stress-induced Cell Death
Scientific Reports (2018)
-
Laboratory Investigation web focus on China
Laboratory Investigation (2016)