Abstract
Epstein–Barr virus (EBV) infection is closely associated with several human malignancies including nasopharyngeal carcinoma (NPC). The EBV immediate-early protein BZLF1 is the key mediator that switches EBV infection from latent to lytic forms. The lytic form of EBV infection has been implicated in human carcinogenesis but its molecular mechanisms remain unclear. BZLF1 has been shown to be a binding partner of several DNA damage response (DDR) proteins. Its functions in host DDR remain unknown. Thus, we explore the effects of BZLF1 on cellular response to DNA damage in NPC cells. We found that expression of BZLF1 impaired the binding between RNF8 and MDC1 (mediator of DNA damage checkpoint 1), which in turn interfered with the localization of RNF8 and 53BP1 to the DNA damage sites. The RNF8-53BP1 pathway is important for repair of DNA double-strand breaks and DNA damage-induced G2/M checkpoint activation. Our results showed that, by impairing DNA damage repair as well as abrogating G2/M checkpoint, BZLF1 induced genomic instability and rendered cells more sensitive to ionizing radiation. Moreover, the blockage of 53BP1 and RNF8 foci formation was recapitulated in EBV-infected cells. Taken together, our study raises the possibility that, by causing mis-localization of important DDR proteins, BZLF1 may function as a link between lytic EBV infection and impaired DNA damage repair, thus contributing to the carcinogenesis of EBV-associated human epithelial malignancies.
Similar content being viewed by others
Main
Epstein–Barr virus (EBV) is a gamma herpesvirus and infection with EBV is associated with a variety of epithelial and B-cell cancers.1, 2 EBV has a biphasic life cycle consisting of latent and lytic stages. The switch from latent to lytic infection is triggered by the EBV immediate-early transcription factor, BZLF1 (ZEBRA, Zta, Z, EB1).3, 4 A number of reports have demonstrated that lytic EBV activation is intimately linked to the pathogenesis of EBV-induced malignancies including the development of NPC.5, 6, 7, 8
The mammalian DNA damage response (DDR) network has pivotal roles in maintaining genome stability and tumor suppression.9, 10, 11, 12, 13 In the presence of double-strand break (DSBs), the protein complex composed of MRE11-RAD50-NBS1 (MRN) recruits and activates the ataxia-telangiectasia-mutated (ATM) kinase at the vicinity of DSBs,10 which leads to the phosphorylation of the histone variant H2AX (γH2AX). γH2AX decorates chromatin domains flanking DSBs, and is directly engaged by the mediator of DNA damage checkpoint 1 (MDC1). MDC1 loading onto the damaged chromatin subsequently permits the productive accumulation of a cohort of DNA damage signaling and mediator proteins.14, 15 In particular, the RING finger ubiquitin ligase RNF8 is targeted to MDC1 via its phospho-binding forkhead-associated domain, where it initiates a cascade of chromatin ubiquitination events important for tethering of DNA damage mediator and repair proteins, including 53BP1 and BRCA1,16, 17, 18, 19 the dysregulation of which compromises genome stability and contributes to cell death and neoplastic transformation.
Interestingly, host DDR proteins have been shown to participate in EBV viral replication.20, 21 Several DDR proteins bind to BZLF1 and those interactions are required for viral replication. DNA damage mediator protein 53BP1 has been reported to interact with BZLF1 via its BRCT domain, although the functional relevance of the complex formation remains obscure.22 53BP1 is intimately involved in the ATM signal-transduction pathway, which is commonly activated during EBV lytic replication. The DNA damage repair protein, Ku80, physically interacts with BZLF1.23 The BZLF1 also binds to and activates p53 (refs. 24 and 25) in certain cell types, which suggests that BZLF1 has a role in inducing DNA damage.26 However, the effects of BZLF1 on DDR are unknown. In this study, we report that BZLF1 suppresses 53BP1 accumulation at DNA damage sites and compromises host cell resistance to genotoxic stress. We have provided evidence that BZLF1 modulates 53BP1 and RNF8 DNA damage foci formation that are involved in DNA damage repair and cell-cycle checkpoint, and that it disrupts the host cellular defense systems for DNA damage.
MATERIALS AND METHODS
Cell Cultures and Transfection
The 293T and HeLa cells were purchased from American Type Culture Collection (ATCC). The DR-U2OS cells were a kind gift from Dr Maria Jasin of Memorial Sloan-Kettering Cancer Center. The cells were cultured in Dulbecco’s modified Eagle’s medium (ATCC) supplemented with 10% fetal bovine serum in 5% CO2 at 37 °C. The HONE1 cells were kindly provided by Dr Ron Glaser (The Ohio State University).27 C666-1 is an EBV-harboring NPC cell line.28 The NPC cell line HK1 was also used in this study.29 HONE1-EBV cells were infected with recombinant Akata EBV that was tagged with GFP according to our previously published protocol.30 HONE1, HONE1-EBV, HK1 and C666-1 cells were cultured in RPMI-1640 medium. For transient transfection, 60% confluent cell cultures were transfected using Fugene HD reagent according to the manufacturer’s instructions (Roche). HONE1-stable cells were generated by retroviral infection. Production of retrovirus was obtained by co-transfection of pVSV-G with pLPCX-BZLF1 or pLPCX into Phoenix 293 packaging cells. HONE1 cells were infected by exposure to retroviral supernatant containing 7 μg/ml polybrene (Sigma) and selected with puromycin (Sigma) for 2 weeks.
Immunofluorescence Analysis
Cells were fixed in 3.7% paraformaldehyde, blocked with 1% bovine serum albumin, permeabilized with 0.5% Triton X-100 and incubated with antibodies against 53BP1 (Bethyl Laboratories), BZLF1 (Argene), RNF8 (Abnova Corporation), γH2AX (Upstate Cell Signaling), MDC1 (Abcam), FK2 (Upstate Cell Signaling), p-ATM (Upstate Cell Signaling) and HA (Sigma) for 1 h. Samples were washed three times with PBS and incubated with secondary antibodies for 1 h. Cells were then stained with Hoechst to visualize nuclear DNA. The coverslips were mounted onto glass slides with antifade solution. Pictures were captured using confocal microscopy (Carl Zeiss LSM 700; Carl Zeiss, Thornwood, NY, USA), under a × 63 objective. For quantification of IR-induced foci (IRIF)-positive cells, about 300 cells were scored for the localization of DDR proteins at IRIF. The cells with ≥10 foci per cell were considered positive for IRIF formation.31
Western Blotting Analysis
Cells were harvested and protein concentrations were determined using Lowry’s assay. Protein was resolved on 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto a PVDF membrane. The membranes were probed with primary antibodies for p-ATM (Epitomics), ATM (Upstate Cell Signaling), p-p53 (Cell Signaling), cleaved-caspase-3 (Cell Signaling), cleaved-PARP (Cell Signaling), HA (Sigma) and Flag (Sigma). Actin (Santa Cruz) was used as a loading control.
Plasmid Construction
pcDNA-BZLF1, Flag-BZLF1 and RFP-BZLF1 were constructed by several subcloning steps. BZLF1 fragments were amplified by polymerase chain reaction (PCR) using BZLF1 expression plasmid p509 (from Dr Wolfgang Hammerschmidt) as a template. The amplified fragments were gel purified, digested (BamHI and ApaI) and cloned into pcDNA3.0 (Invitrogen). The PCR product was subcloned into the pcDNA3.0 (Invitrogen) using the primers 5′-ATATGGATCCATGATGGACCCAAACT-3′ and 5′-AGATGAATTCTTAGAAATTTAAGAGA-3′. The obtained plasmid was sent to BGI Health (Hong Kong) for sequencing. pcDNA3.0 was transfected in parallel as a control. Similarly, BZLF1 fragments were amplified by PCR using BZLF1 expression plasmid p509. The PCR product was digested with BamHI and EcoRI restriction enzymes. The digested PCR product was subcloned into the RFP vector. The primers used were: 5′-ATATGGATCCATGATGGACCCAAACT-3′ and 5′-AGATGAATTCGAATTTAAGAGATCC-3′. Flag tag was placed at the N terminus of BZLF1 by PCR amplification and subcloned into pcDNA 3.0 vector. The PCR primers used were 5′-ATATGGATCCGACTACAAAGACGATGACGATAAAATGATGGACCCAAACT-3′ and 5′-ATATGGGCCCTTAGAAATTTAAGAGA-3′. The sequence of the generated plasmid was confirmed by BGI Health (Hong Kong). The pLPCX-BZLF1 retrovirus vector was constructed by subcloning BZLF1 into pLPCX vector at BamHI and XhoI restriction sites.
Pull-Down Assay
For immunoprecipitation, cells were harvested with NETN buffer (0.5% NP-40, 2 mm EDTA, 50 mm Tris-Cl, pH 8.0, and 100 mm NaCl) after treatment and washed once with PBS. After centrifugation, lysates were incubated with protein A-agarose beads (GE Health Life Sciences) together with antibodies against Flag (Sigma), HA (Santa Cruz) or 53BP1 at 4 °C overnight with gentle agitation. For streptavidin bead pull-down assay, the lysates were incubated for 4 h at 4 °C with streptavidin beads (Invitrogen). Beads were washed three times with NETN buffer. The protein complexes immobilized on the beads were subjected to western blotting.
Cell Fractionation Experiments
To assess subcellular localization of RNF8, 293T cells transfected with Flag-BZLF1 or control vector were lysed with NETN buffer for 10 min on ice. Lysates were centrifuged, and the supernatant (soluble fraction) was decanted. The resulting pellet, enriched in chromatin-bound proteins, was sonicated and boiled in sampling buffer.
Clonogenic Survival Assay
Approximately 2000 cells were seeded onto 35 mm dish in triplicates. Twenty-four hours after seeding, cells were exposed with different doses of IR. The medium was replaced 24 h later and cells were then incubated for 7 days. Resulting colonies were fixed and stained with 2% crystal violet. Colonies were counted using a GelDoc with Quantity One software (Bio-Rad). Results were presented as the averages of data obtained from three independent experiments.
G2/M Cell-Cycle Checkpoint Assay
HONE1 cells were transfected with control or BZLF1 plasmid. Twenty-four hours after transfection, transfected cells were irradiated at 6 Gy. Twenty-four hours after irradiation, cells were fixed with 70% (v/v) ethanol at 4 °C for 24 h, then incubated with p-H3 antibody (Cell Signaling) for 1 h, followed by secondary antibody for another 1 h. The stained cells were treated with RNase A, incubated with propidium iodide and then analyzed by flow cytometry.
Homologous Recombination Repair Assay
Cells were co-transfected with BZLF1 or pcDNA together with pCBA-SceI plasmid into DR-GFP U2OS cells. The homologous recombination (HR) repair assay was performed according to the previous report.32 Cells were plated in 35 mm dishes and incubated in culture media for 48 h before fluorescence-activated cell sorting (FACS) analyses. Results were the averages of data obtained from three independent experiments.
Non-Homologous End-Joining Assay
DNA repair by non-homologous end-joining (NHEJ) assay was assessed by a random plasmid integration assay.33 HONE1-stable cells were transfected with pcDNA3.1/hygro plasmid linearized by BamHI and XhoI digestion (Roche) plus the pEGFP-C1 plasmid. Twenty-four hours after transfection, cells were plated into two separate plates. The following day, one plate of cells was fixed and examined under fluorescence microscope to determine the transfection efficiency by EGFP expression. The other plate was maintained in selective media containing 50 μg/ml hygromycin for 7 days at 37 °C to allow for colony formation. The colony is defined to consist of at least 50 cells. Colonies were stained with 2% crystal violet and were quantified with Quantity One software (Bio-Rad). The random plasmid integration efficiency was normalized for transfection and plating efficiencies.
Spectral Karyotyping Analysis
Metaphase chromosome spreads were prepared on specially treated glass slides. The slide was treated with DNase-free RNase solution (0.1 mg/ml) at 37 °C for 1 h, washed in 2 × SSC (sodium chloride and sodium citrate) solution for 10 min at room temperature (RT), dehydrated in 70, 85 and 95% ethanol at RT for 2 min and air dried. Next, the slide was treated with proteinase K (0.05 μg/ml) at 37 °C, washed in 2 × SSC at RT, fixed in 2% paraformaldehyde and washed in 2 × SSC two times at RT, for 10 min each, followed by dehydration in ethanol as above. The slide was placed in 70% formamide solution at 70 °C for 4 min, and dehydrated in ethanol. Then, 4 μl of spectral karyotyping (SKY) probe from Applied Spectral Imaging (ASI, Migdal Ha’Emek, Israel) was denatured at 80 °C for 7 min. The denatured probe was incubated at 37 °C for 1 h before adding onto the slide. The probed slide was covered by a coverslip, sealed and incubated in a humidified chamber at 37 °C for about 36 h. The detection procedures followed the recommendations of ASI. SKY images were captured using the SkyVision Imaging System equipped with a Zeiss Axioplan 2 fluorescence microscope. Karyotyping was performed using the special software provided by ASI (SKY View 2.0).
MTT Assay
Cell number was monitored using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay. Briefly, C666-1 cells were seeded into 96-well plates and incubated overnight before transfection with pcDNA or BZLF1 plasmid. At 24 h after transfection, cells were treated with camptothecin. At 24, 48 and 72 h after treatment, MTT was added to each well. After 4 h incubation, the absorbance of each well was measured at 570 nm. All experiments were performed in triplicate.
RESULTS
BZLF1 Blocks the Formation of 53BP1 Foci
One of the key downstream steps of DSB response is the recruitment of 53BP1 to DSB-flanking chromatin.34 Following exposure to IR, 53BP1 is rapidly recruited to DSB sites to form IRIF. It has been reported that the EBV lytic gene, BZLF1, binds to 53BP1, and that knockdown of 53BP1 reduces the efficiency of EBV lytic replication.22 We examined if BZLF1 might affect the function of 53BP1. We first examined if recruitment of 53BP1 to IRIF may be affected in BZLF1-expressing cells. This part of the study was performed using HONE1 cells. As seen in Figure 1a, expression of BZLF1 in HONE1 cells blocked the formation of 53BP1 foci, whereas expression of vector control pcDNA in parallel experiments did not interfere with the recruitment of 53BP1 to DSBs (Figure 1a). Expression of EGFP as an additional control did not displace the 53BP1 from IRIF, which further supported that the BZLF1-induced mislocalization of 53BP1 was specific (Figure 1b). These consistent results suggest that BZLF1 can block 53BP1 recruitment to IRIF.
BZLF1 blocks the formation of 53BP1 foci. (a) HONE1 cells were transfected with control pcDNA or BZLF1 vector and treated with 6 Gy ionizing radiation (IR). Cells were fixed 3 h after IR, stained for BZLF1 and 53BP1 antibodies. Nuclei were stained with Hoechst. Transfected HONE1 cells (300) were scored for the localization of 53BP1 at IR-induced foci (IRIF). Data are represented as mean±s.d. *P<0.05, BZLF1-transfected group compared with pcDNA-transfected cells. (b) HONE1 cells transfected with EGFP vector were subjected to immunofluorescence (IF) microscopy 3 h after irradiation (6 Gy). Cells were stained for 53BP1 and merged with Hoechst stain of DNA. (c) Co-immunoprecipitation experiments to detect interaction between 53BP1 and BZLF1. Protein extract from 293T cells transfected with Flag-BZLF1 was subjected to immunoprecipitation with anti-Flag or anti-53BP1 antibodies. (d) The 293T cells were co-transfected with plasmids encoding HA-tagged wild-type or mutants of 53BP1 lacking BRCT domain and Flag-BZLF1. Immunoprecipitation was performed using anti-Flag antibody and co-precipitating HA-53BP1 was detected by immunoblotting. (e) HONE1 cells were co-transfected with HA-tagged wild-type or its deletion mutant of 53BP1 and pcDNA/BZLF1 plasmids. Cells were irradiated 24 h after transfection, and then immunostained using anti-HA and BZLF1 antibodies. About 300 cells were scored for the localization of 53BP1 at IRIF. Data are represented as mean±s.d. *P<0.05, BZLF1-transfected group compared with pcDNA-transfected cells.
There are at least two possible mechanisms by which BZLF1 interferes with 53BP1 recruitment to IRIF: either BZLF1 affects the upstream regulators involved in 53BP1 foci formation or BZLF1 directly competes with 53BP1 for binding to the damage-modified chromatin. We performed co-immunoprecipitation experiment to confirm the interaction between 53BP1 and BZLF1. As shown in Figure 1c, 53BP1 protein co-precipitated with Flag-BZLF1. The reciprocal immunoprecipitation was performed using anti-53BP1 antibody and Flag-BZLF1 was pulled down. The co-immunoprecipitation results suggest that BZLF1 is present in the 53BP1 complex. It was previously reported that the BRCT domain of 53BP1 is responsible for BZLF1 binding.22 To determine whether 53BP1 interacts with BZLF1 through BRCT tandem repeats, we performed immunoprecipitation experiment in cells that were co-transfected with plasmids expressing Flag-BZLF1 and HA-53BP1 or HA-53BP1 without the BRCT domain (HA-53BP1ΔBRCT).35 The results showed that Flag antibodies could only pull down HA-53BP1 but not HA-53BP1ΔBRCT (Figure 1d), which confirmed that 53BP1 associates with BZLF1 through its BRCT domain. To further discriminate between the two possible mechanisms by which BZLF1 interferes with the recruitment of 53BP1 to IRIF, HONE1 cells were transfected with expression plasmids encoding HA-53BP1ΔBRCT and then assayed for 53BP1 recruitment at DNA damage sites following IR. As with the wild-type 53BP1, the expression of BZLF1 prevented the recruitment of mutant 53BP1 to IRIF. The percentage of BZLF1-expressing cells showing this phenotype was similar to that of cells expressing wild-type 53BP1 (Figure 1e). This is consistent with a previous study, which reports that 53BP1 recruits to DSBs through its tandem Tudor domain but not BRCT domain.35 Our results therefore do not support a role of BZLF1 as a competitor of 53BP1 for binding to chromatin.
BZLF1 Attenuates the Formation of FK2 and RNF8 Foci but not p-ATM, MDC1 and γH2AX Foci
Next, we examined if the upstream regulators of 53BP1 may be affected by expression of BZLF1. Localization of 53BP1 to IRIF requires the accumulation of p-ATM, γH2AX and MDC1 at DSB sites. DSB is recognized by MRN followed by the activation of ATM, which phosphorylates the H2AX histone to form γH2AX. The p-ATM and γH2AX are accumulated onto IRIFs upon IR. We analyzed the formation of p-ATM and γH2AX foci as the early stage of DDR in BZLF1-expressing and control cells. In addition, we also assessed the recruitment of MDC1 to IRIFs after expression of BZLF1. Our results showed that p-ATM, γH2AX foci formation and the recruitment of MDC1 to the sites of DNA damage were not affected after expression of BZLF1 (Figures 2a–c). We then examined if BZLF1 affects DSB ubiquitination by performing immunostaining experiments using the FK2 antibody, which recognizes mono- and polyubiquitinated proteins but not free ubiquitin. Our results showed the IR-induced accumulation of ubiquitinated substrates, as detected by the FK2 antibody, was abrogated after expression of BZLF1 (Figure 2d). Without ionizing radiation (IR) treatment, the single 53BP1 nuclear focus staining pattern was consistent with that of 53BP1 nuclear bodies reported previously.36 Overall, we conclude from these results that expression of BZLF1 is dispensable for the upstream events of p-ATM foci formation. Accumulation of γH2AX and recruitment of MDC1 to the damage sites, which depend on ATM-mediated phosphorylation but not on ubiquitination, are not affected by BZLF1 expression. As RNF8 acts with UBC13 in promoting protein ubiquitination at or near DNA damage sites,16, 18 we then examined RNF8 foci formation in BZLF1-expressing cells. In line with the abrogation of ubiquitinated substrate accumulation by BZLF1, RNF8 foci were absent in these BZLF1-expressing cells (Figure 2e). We therefore conclude that BZLF1 compromises RNF8 docking at DSB and attenuates IR-induced ubiquitination.
BZLF1 attenuates the formation of FK2 and RNF8 foci but not p-ATM, MDC1 and γH2AX foci. (a) Immunofluorescence (IF) staining for γH2AX was performed in HONE1 cells transfected with either pcDNA or BZLF1 plasmid and exposed to irradiation (6 Gy) at 24 h after transfection. Hoechst was used to counterstain the nucleus. Representative IF images are shown. (b) IF double staining of p-ATM (green) and BZLF1 (red) in HONE1 cells transfected with pcDNA or BZLF1 plasmid, and then analyzed 3 h after irradiation with 6 Gy. DNA was counterstained with Hoechst (blue), and merged images were also shown. (c) HONE1 cells transfected with pcDNA or BZLF1 plasmid were analyzed by IF with 53BP1 and MDC1 antibodies. Cells were treated with 6 Gy ionizing radiation (IR) and analyzed by IF 3 h after IR. Quantification of (a)–(c) is shown. 53BP1, MDC1, p-ATM and γH2AX IR-induced foci (IRIF) were counted and graphed. About 300 cells were scored for the localization of those proteins at IRIF in each group. Data are represented as mean±s.d. *P<0.05, BZLF1-transfected group compared with pcDNA-transfected group. For (d) and (e), the experiments were carried out in the same manner as in (a)–(c), except using indicated antibodies, and were also performed in cells without IR treatment. Quantification of 53BP1 IRIF was obtained from 300 damaged cells in each group from two independent experiments. Data are represented as mean±s.d. *P<0.05, BZLF1-transfected group compared with pcDNA-transfected group.
BZLF1 Impairs the Binding Between MDC1 and RNF8
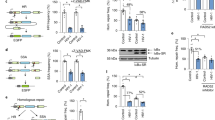
The RNF8 protein contains a forkhead-associated domain that binds ATM-catalyzed phospho-TQXF motifs on MDC1.16 This binding is required for its localization to DNA damage foci and further accumulation of 53BP1 to IRIF.16, 17 Therefore, it is conceivable that absence of RNF8 foci formation might be caused by the disruption of the binding between MDC1 and RNF8. To examine this possibility, we co-transfected cells with pcDNA/BZLF1, HA-MDC1 and SFB (S, Flag and streptavidin-binding protein)-tagged RNF8 constructs, and then examined if BZLF1 interferes with the protein interaction between MDC1 and RNF8 using immunoprecipitation assay. Indeed, a lower level of MDC1 was pulled down by RNF8 in BZLF1-expressing cells with and without IR (Figure 3a). To further verify if BZLF1 interacts with RNF8, we coexpressed HA-tagged MDC1, BZLF1 and SFB-RNF8 in 293T cells, and then the cell lysates were subjected to pull down with streptavidin beads, which bind to the SFB-RNF8. BZLF1 was readily detectable in the precipitate (Figure 3b). This suggests that BZLF1 can interact with RNF8. To confirm whether BZLF1 interacts with RNF8, we transfected cells with SFB-RNF8 and BZLF1 plasmids, and used anti-Flag antibody to perform immunoprecipitation assay. BZLF1 protein was shown to co-precipitate with RNF8, thus demonstrating that BZLF1 interacts with RNF8 (Figure 3c).
BZLF1 impairs the binding between MDC1 and RNF8. (a) The 293T cells were co-transfected with HA-MDC1, SFB-RNF8 and pcDNA or BZLF1 plasmids. Twenty-four hours after transfection, cells were treated with 6 Gy ionizing radiation (IR) and harvested. Lysates were incubated with anti-Flag antibody or anti-IgG antibody together with protein A sepharose beads for 4 h at 4 °C. Thereafter, beads were washed three times with NETN (20 mM Tris, pH 8, 100 mM NaCl, 1 mM EDTA, 0.2% Nonidet P-40) buffer, isolates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed by western blotting using indicated antibodies. (b) The 293T cells were transfected with plasmids encoding SFB-RNF8 along with those encoding HA-MDC1 and pcDNA or BZLF1. Precipitation was conducted using streptavidin beads and immunoblotting was performed using anti-Flag, anti-BZLF1 or anti-HA antibodies as indicated. (c) The 293T cells were co-transfected with SFB-RNF8 along with BZLF1 plasmid. Immunoprecipitation was carried out using anti-Flag antibody. Immunoblotting was performed using anti-Flag or anti-BZLF1 antibodies. (d) Chromatin lysates or total cell extracts were prepared from pcDNA- or BZLF1-transfected cells with or without IR treatment. Immunoblotting was conducted using anti-RNF8, anti-MDC1 and anti-BZLF1 antibodies.
To further confirm that BZLF1 impairs DSB localization of RNF8, we performed subcellular fractionation experiments and determined the binding of RNF8 and MDC1 proteins on chromatin in the presence or absence of BZLF1 expression. There was no difference in the amount of MDC1 recruitment on chromatin between pcDNA- and BZLF1-expressing cells. However, there was a significant decrease of RNF8 loading on chromatin in cells with BZLF1 expression, despite the fact that IR treatment induced RNF8 recruitment on chromatin in control cells (Figure 3d). By contrast, the RNF8 level in the whole-cell lysates did not change after DNA damage (Figure 3d). All these results show that BZLF1 impairs the binding between RNF8 and MDC1, and the accumulation of RNF8 onto chromatin.
BZLF1 Impairs DSB Repair and Activates ATM-Dependent Signaling Pathways
Given that recruitment of 53BP1 is required for efficient DSB repair, our results raised the possibility that expression of BZLF1 may delay or prevent repair of IR-induced DNA damage. To test this hypothesis, DNA repair capacity was monitored by assessing γH2AX foci formation, which served as a marker of DNA damage. We irradiated the stable BZLF1-expressing cells with a dose of 2 Gy and counted the γH2AX foci numbers at several time points. As expected, γH2AX rapidly decreased in control vector-infected cells, consistent with ongoing repair of DNA damage after IR. By contrast, the γH2AX signal decreased at a much slower rate in the majority of the irradiated BZLF1-expressing cells, particularly during the later time points (Figure 4a). Overall, these data support the conclusion that BZLF1 impairs DSB repair. DNA DSBs are repaired by two major systems: NHEJ and HR. NHEJ is an intrinsically error-prone repair system that operates throughout the cell cycle. HR is an error-free repair system, but it is limited to the late S and G2 phases because the replicated DNA strand is used as a template.37 We used a GFP-based repair assay for HR repair in DR-GFP U2OS cells as described previously.38 U2OS cells with a single integrated copy of the HR repair substrate were co-transfected with I-SceI and pcDNA or BZLF1 plasmids, followed by FACS analysis of GFP-positive events to assay for DSB repair by HR. HR was slightly reduced (Figure 4b). The 53BP1 has a major role in NHEJ repair.39 RNF8 also facilitates NHEJ repair by regulating the abundance of the NHEJ repair protein KU80 at sites of DNA damage.40 We examined NHEJ repair by using a well-characterized plasmid-based end-joining assay. In brief, HONE1 BZLF1-stable cells were transfected with pcDNA3.1/hygrolinearized by BamHI and XhoI digestion. Cells were selected with hygromycin for 7 days, and DNA damage repair via NHEJ was measured by relative colony number formation. Consistent with the requirement of 53BP1 and RNF8 for NHEJ repair, BZLF1-expressing cells showed a 2.5-fold reduction in NHEJ repair capability (Figure 4c).
BZLF1 impairs double-strand break (DSB) repair and activates ataxia-telangiectasia mutated (ATM)-dependent signaling pathways. (a) Quantification of γH2AX foci in pLPCX- and pLPCX-BZLF1-stable HONE1 cells over a period of 24 h after irradiation with doses of 2 Gy. (b) The homologous recombination (HR) repair efficiency was determined by analyzing the percentage of GFP-positive cells. Data are presented as mean±s.d. (n=3). (c) The non-homologous end-joining (NHEJ) efficiencies in pLPCX- or pLPCX-BZLF1-expressing cells were determined, and relative NHEJ efficiencies were calculated by normalizing to control pLPCX groups. The data are presented as mean±s.d. (n=3). *P<0.05, pLPCX-BZLF1 group compared with pLPCX control group. (d) BZLF1 cells showed an activation of ATM-dependent signaling pathways. Western blot analysis of DDR pathway-related protein levels in whole-cell extracts from HONE1 cells transfected with pcDNA or BZLF1 plasmid. Twenty-four hours after transfection, cells were treated with 6 Gy ionizing radiation (IR). Extracts were prepared at the times indicated following exposure to IR.
The ATM signaling is activated following DNA damage.41 The accumulation of DNA damage arising from a compromised repair may activate ATM signaling pathway. To investigate the status of ATM signaling pathway in BZLF1-expressing cells, a time-course study of key events over a period of 24 h after exposure to 6 Gy of IR was carried out (Figure 4d). Interestingly, p-ATM (ser-1981), which is regarded as a marker of ATM activation,42 showed an increased signal in BZLF1-expressing cells compared with a control cell line at the same time point, and the increase was sustained over a 24 h time period. P53 is a well-known downstream target of ATM and ATM phosphorylates p53 on serine 15.43 Ectopic expression of BZLF1 caused an increase in p-p53 level in BZLF1-expressing cells, compared with pcDNA control cells. Another substrate phosphorylated by ATM is histone H2AX.44 Expression of phosphorylated H2AX (γH2AX) was also increased in the BZLF1-expressing cells. In contrast to these markers of DNA damage-induced signaling, the levels of the 53BP1, MDC1 and RNF8 were not significantly affected in BZLF1-expressing cells with or without IR treatment. These data demonstrate that BZLF1 expression results in sustained DNA damage signaling.
BZLF1 Abrogates the G2/M Checkpoint and Increases Chromosomal Structural Aberrations
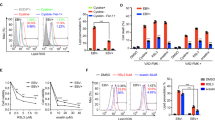
It is well documented that ATM and ATM-dependent signaling pathways are primarily responsible for controlling the activation and maintenance of DNA damage-induced cell-cycle checkpoints after exposure to IR.43 The 53BP1 itself can functionally regulate G2/M-phase DNA damage-activated cell-cycle checkpoints after IR.45, 46 BZLF1 induces cell-cycle arrest in different cell lines,47, 48 and both G2 and mitotic block in HeLa cells.49 As our study showed that BZLF1 could block the accumulation of 53BP1 and RNF8 proteins at the sites of DNA breaks, we further examined the effect of BZLF1 on IR-induced cell-cycle arrest. The effect of BZLF1 on activating the G2/M checkpoint was first examined in HONE1 cells. BZLF1-expressing and pcDNA control cells were irradiated with 6 Gy of IR and harvested after 24 h. The cells were fixed and stained with an antibody directed against phosphorylated serine-10 of histone H3, which is a well-documented marker of G2/M cells,50 and subjected to flow cytometric analysis. A decrease in the percentage of mitotic cells was observed in control cells after irradiation, indicating that the G2/M checkpoint was functional in these cells. By contrast, a higher percentage of BZLF1-expressing cells entered mitosis 24 h after irradiation, which demonstrates that BZLF1 can indeed abrogate G2/M checkpoint (Figure 5a).
BZLF1 abrogates the G2/M checkpoint and increases radiosensitivity of cells. (a) G2/M checkpoint response was verified by labeling mitotic cells using an anti-phospho-H3 antibody in ionizing radiation (IR)-treated HONE1 cells transfected with pcDNA or BZLF1 plasmid. At the same time, cells were stained with propidium iodide for cell-cycle analysis. Bar graph shown is the average of three experiments. *P<0.05, BZLF1-transfected group compared with corresponding pcDNA-transfected cells. (b) Cells transfected with pcDNA or BZLF1 plasmid were treated with increasing doses of IR, and then incubated for 7 days to allow the growth of colonies for the clonogenic survival assay. Data points represent the mean±s.d. of triplicate determinates of three independent survival experiments.
We also examined the radiosensitivity of cells after expression of BZLF1. BZLF1-expressing cells exhibited a moderate but reproducible hypersensitivity to IR, as measured by colony formation assay (Figure 5b), which further supports that BZLF1 has a role in cellular response to DNA damage. The defective DNA damage signaling pathway and DNA damage repair ability in BZLF1-expressing cells suggest that BZLF1 expression may induce genomic instability in cells. To examine if BZLF1 induces chromosomal aberrations, we transfected HeLa and HONE1 cells with BZLF1 or control vector for 24 h and subjected the cells to karyotype analysis using 24-color SKY. Chromosomal structural aberrations, including chromatid breaks or deletions, chromosome breaks, chromosomal rearrangements, chromosome rings, isochromosomes and chromosome duplications were examined. Cells transfected with BZLF1 were shown to have significantly (P<0.05) higher frequencies of total non-clonal chromosome structural aberrations than the vector-transfected control cells (Supplementary Figure 1, Supplementary Figure 2 and Table 1). The frequencies of non-clonal numerical aberrations did not show significant differences between BZLF1-transfetced or control vector-transfected cells (data not shown). The results of chromosome aberration analysis show that BZLF1 has the ability to induce chromosomal structural instability.
Micronucleus assay is a method that is commonly used to measure chromosome damage or genomic instability in cells. The number of micronuclei formation is closely correlated with the extent of DNA damage. It is a reliable method for measuring chromosome breaks or chromosome loss.51, 52 To examine the ability of BZLF1 to induce micronuclei formation in human epithelial cells, the micronuclei formation in HONE1 cells was examined after transfection with RFP-BZLF1 or control RFP plasmid. Cells showed that a micronuclei phenotype was enumerated. The results demonstrated that there were almost twofold more RFP-BZLF1-expressing cells than the RFP control cells that exhibited this phenotype (Supplementary Figure 3). Hence, expression of BZLF1 increases chromosomal aberrations.
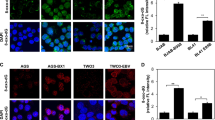
BZLF1 Blocks Formation of 53BP1 and RNF8 Foci in EBV-Positive Cells and Sensitizes Cells to Chemotherapeutic Drugs
To explore the physiological relevance of BZLF1 to DDR, we next examined role of BZLF1 on host DDR in the context of EBV-infected cells. EBV lytic reactivation, including BZLF1 expression, can be chemically induced in EBV-infected HONE1 cells. We first confirmed that expression of BZLF1 but not the control vector could block formation of 53BP1 foci after IR in HONE1-EBV cells (Figure 6a). Those cells without 53BP1 foci were BZLF1 positive but not necessarily EBV positive. We then induced EBV lytic reactivation in HONE1-EBV cells using 12-O-tetradecanoylphorbol-13-acetate (TPA) plus sodium butyrate. BZLF1 expression was induced during lytic reactivation. Blockage of 53BP1 foci was observed in BZLF1-positive cells after EBV lytic reactivation (Figure 6a). MDC1 foci formation in Figure 6b remained intact. As in HONE1 cells, DNA damage-induced ubiquitination in HONE1-EBV cells after expression of BZLF1, as indicated by FK2 and RNF8 staining, was abrogated (Figures 6c and d).
BZLF1 blocks the formation of 53BP1 and RNF8 foci in Epstein–Barr virus (EBV)-positive cells and sensitizes cells to chemotherapeutic drugs. (a) HONE1-EBV cells were transfected with either pcDNA or BZLF1 plasmid or treated with 12-O-tetradecanoylphorbol-13-acetate (TPA) plus sodium butyrate (SB). After twenty-four hours, cells were treated with 6 Gy IR and recovered for 3 h before they were fixed and permeabilized. Immunostaining experiments were performed using 53BP1 and BZLF1 antibodies. (b–d) The experiment was carried out in the same manner as in (a), except using indicated antibodies. (e) C666-1 cells were transfected with pcDNA or BZLF1 plasmid and cells were treated with camptothecin. Cell division rates were monitored using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay 24, 48 and 72 h after adding the drug. MTT assays were performed in triplicate, and the mean absorbance values at 570 nm±s.d. are presented. (f) Western blotting analysis of cleaved-caspase-3 and cleaved-PARP in whole-cell lysates of pcDNA- or BZLF1-transfected HONE1-EBV cells treated with adriamycin or cells were pretreated with 120 μg/ml phosphonoacetic acid (PAA) for 30 min before adding adriamycin to the cells.
As BZLF1 expression could interfere with DNA damage repair by impairing the recruitment of 53BP1 to IRIF, we postulated that BZLF1 expression might sensitize NPC cells to chemotherapeutic drugs. We first determined the effect of camptothecin treatment on the proliferation rate of EBV-positive NPC cell line C666-1 expressing pcDNA or BZLF1 using MTT assay. Figure 6e showed that the cytotoxic effect of camptothecin was more pronounced in EBV-positive cells transfected with BZLF1, compared with vector control cells. HONE1-EBV cells were also used to compare the toxicity of another chemotherapy drug, adriamycin. Western blot analysis showed that BZLF1 caused increased apoptosis in HONE1-EBV cells with or without adriamycin treatment, as indicated by the increased level of cleaved-caspase 3 and cleaved-PARP. To exclude the possibility that the increased apoptosis was induced by the viral production, we also pretreated the cells with phosphonoacetic acid (PAA), an inhibitor of viral DNA polymerase, before adriamycin treatment. PAA did not block the adriamycin-induced apoptosis in BZLF1-expressing cells (Figure 6f). Taken together, our results indicate that BZLF1 blocks 53BP1, as well as FK2 and RNF8 foci formation upon IR, and consequently renders EBV-positive cells more sensitive to chemotherapeutic drugs.
DISCUSSION
The function of BZLF1 as a transcriptional factor is well established. BZLF1 shares structural similarities to the basic leucine zipper family of transcriptional factors. It is involved in the activation of the replication origin of EBV, oriLyt, and is crucial in the activation of lytic EBV infection in cells. In this current study, we report a novel function of EBV immediate-early protein BZLF1 in DDR pathways in its host cells. We found that BZLF1 impairs recruitment of 53BP1 and RNF8 to DSBs, which may interfere with the cellular activity to repair DNA damage, including activation of the G2/M checkpoint,45, 46 as well as repair of DNA DSBs via NHEJ.53 Thus, we propose that BZLF1 can impair DNA damage repair and G2/M checkpoint. The functional studies showed that expression of BZLF1 not only sensitized cells to IR but also induced genomic instability in cells. The molecular mechanisms involved in this process were also studied, and we found that BZLF1 competes with MDC1 for binding to RNF8, and in effect displaces RNF8 from DSB and impairs the local recruitment of DNA damage mediator protein 53BP1 to DNA damage sites. Moreover, BZLF1-induced blockage of 53BP1, FK2 and RNF8 foci formation is not cell-type-specific. When BZLF1 was transfected into HK1 cells, it also interfered with the formation of 53BP1, FK2 and RNF8 foci, as shown in Supplementary Figures 4 and 5. Hence, BZLF1-induced phenotype could be recapitulated in NPC cells.
EBV infection is present in almost all undifferentiated NPCs and has been postulated to be an important etiological agent. Lytic activation may contribute to genomic instability and have a role in NPC pathogenesis. Furthermore, radiotherapy and chemotherapy are the main treatment options; both of them induce DNA damages. NPC is particularly sensitive to radiotherapy. The underlying mechanism of this sensitivity is not completely understood. Cells respond to DNA damage by activating cell-cycle checkpoint and repair mechanisms. If the DNA damage is unrepaired, cells will trigger the apoptosis pathway. The impairment of DNA DSB repair correlates with cell radiosensitivity to killing, and thus, the abrogation of G2/M checkpoint and inhibition the DNA DSB repair by BZLF1 may have a role in enhancing the sensitivity of EBV-infected NPC cells to DNA damage-induced apoptosis. Recent studies show that EBV induces genomic instability by inducing DNA damage as well as inhibiting damage repair, but the mechanisms involved remain controversial.7, 54 In this study, we observed that BZLF1 could induce genomic instability. BZLF1 inhibited NHEJ DSBs repair, and the deregulation of the G2/M checkpoint, both of which may contribute to genomic instability.
With respect to the two important DSB repair pathways, BZLF1 primarily impairs NHEJ repair and only has slight effect on HR repair. This observation is consistent with previous linkage between 53BP1 and NHEJ that 53BP1 mainly functions in NHEJ repair pathway.55, 56 How BZLF1 disrupts NHEJ is not clear at this stage. BZLF1 has been reported to bind to Ku80 through its C-terminal region.23 This interaction may also prevent the Ku70/Ku80 proteins from binding to DNA DSB ends to serve as a scaffold for subsequent NHEJ repair reactions. Further work will be required to dissect the potential relationships among 53BP1, Ku80 and BZLF1 during NHEJ-mediated repair.
Expression of EBV lytic proteins was previously found to inhibit apoptosis in EBV-infected B cells such as Burkitt lymphoma-derived cells.57, 58, 59 Those reports showed that BZLF1-positive cells are negative for TUNEL (terminal deoxynucleotidyl transferase dUTP nick-end labeling) staining and suppression of lytic cycle in B cells can induce more apoptosis in cells. On the contrary, we have demonstrated that expression of BZLF1 in EBV-positive NPC cells enhanced apoptosis, which is consistent with what was previously described for epithelial cells in which viral lytic cycle induces EBV-related cell death.60 Thus, the cell death events following viral lytic cycle induction in B cells and epithelial cells could be totally opposite.
DDR signaling could have detrimental effects on the viral life cycle. Once virus enters the host cells, the host cell often responds with defense mechanisms such as DNA damage repair. This is threatening to viruses and therefore viruses take control of the repair signaling pathways for its own benefit.61 The recruitment of DDR proteins to damaged sites is crucial for DNA damage repair. It is common for viral proteins to target directly cellular repair pathways by degrading or causing mislocalization of those key players in the cellular defense pathway. For example, the herpes simplex virus-1-encoded E3 ubiquitin ligase ICP0 binds to RNF8 via the RNF8 FHA domain, and degrades RNF8 and RNF168.62 Human cytomegalovirus avoids the activation of the DNA damage checkpoint by mislocalization of checkpoint proteins including ATM and Chk2.63 In Kaposi’s sarcoma-associated herpesvirus, vIRF1 protein binds to the cellular ATM kinase to prevent DDR.64 Adenovirus prevents the DDR by targeting the MRN complex for degradation and mislocalization.65, 66 Therefore, viruses and virus-encoded proteins can bind to and limit the function of key DNA damage signal transducers. Our data presented here reveal a novel function of EBV-encoded lytic gene. BZLF1 does not degrade DDR proteins; instead, it impairs the binding between two important DDR proteins and leads to mislocalization of 53BP1 and RNF8. In our study, the ATM signaling pathway is still activated. This outcome of the signaling pathway is beneficial for EBV lytic cycle. It has been reported that ATM activity is required for the EBV reactivation.67 Wild-type p53 is also crucial for EBV lytic cycle. Here, we show that BZLF1 activates ATM. BZLF1-induced activation of ATM can transduce to its downstream target p53. As a result, activated ATM and p53 contribute to further activation of EBV lytic cycle with the expression of BZLF1 protein. Thus, the activation of ATM by BZLF1 establishes a positive feedback loop that promotes EBV lytic cycle. The BZLF1-induced activation of ATM is beneficial for EBV lytic cycle induction.
EBV infection is associated with the development of various human cancers. Recently, more and more EBV genes have been found to have functions in DDR. It has long been assumed that latent genes products contribute to EBV-induced tumorigenesis. LMP1 represses DNA damage repair through inactivation of FOXO3a.68, 69 EBNA1 induces DNA damage through the production of reactive oxygen species.70 EBNA3C disrupts cell-cycle checkpoint and has oncogenic activity.71 The lytic EBV infection and the expression of lytic genes may have direct impact on genomic instability. The EBV-encoded Ser/Thr kinase BGLF4 induces a DNA damage signal and induces chromosomal abnormality.72 BZLF1 has been indicated in inducing DNA damage,26 but the exact mechanism is obscure. In this study, we have demonstrated that BZLF1 impairs localization of DDR proteins 53BP1 and RNF8 to IRIF, which may have impact on genomic instability in EBV-infected cells. Our data support that lytic EBV infection has a role in promoting human cancer, particularly epithelial-derived carcinoma in which EBV infection is mainly lytic in nature.
References
zur Hausen H, Schulte-Holthausen H, Klein G et al. EBV DNA in biopsies of Burkitt tumours and anaplastic carcinomas of the nasopharynx. Nature 1970;228:1056–1058.
Cohen JI . Epstein–Barr virus infection. N Engl J Med 2000;343:481–492.
Countryman J, Miller G . Activation of expression of latent Epstein–Barr herpesvirus after gene transfer with a small cloned subfragment of heterogeneous viral DNA. Proc Natl Acad Sci USA 1985;82:4085–4089.
Chevallier-Greco A, Manet E, Chavrier P et al. Both Epstein–Barr virus (EBV)-encoded trans-acting factors, EB1 and EB2, are required to activate transcription from an EBV early promoter. EMBO J 1986;5:3243–3249.
Hirayama T, Ito Y . A new view of the etiology of nasopharyngeal carcinoma. Prev Med 1981;10:614–622.
Hong GK, Gulley ML, Feng WH et al. Epstein–Barr virus lytic infection contributes to lymphoproliferative disease in a SCID mouse model. J Virol 2005;79:13993–14003.
Fang CY, Lee CH, Wu CC et al. Recurrent chemical reactivations of EBV promotes genome instability and enhances tumor progression of nasopharyngeal carcinoma cells. Int J Cancer 2009;124:2016–2025.
Huang SY, Fang CY, Tsai CH et al. N-methyl-N'-nitro-N-nitrosoguanidine induces and cooperates with 12-O-tetradecanoylphorbol-1,3-acetate/sodium butyrate to enhance Epstein–Barr virus reactivation and genome instability in nasopharyngeal carcinoma cells. Chem Biol Interact 2010;188:623–634.
Lou Z, Chen J . Mammalian DNA damage response pathway. Adv Exp Med Biol 2005;570:425–455.
Jackson SP, Bartek J . The DNA-damage response in human biology and disease. Nature 2009;461:1071–1078.
Bartek J, Lukas J . DNA damage checkpoints: from initiation to recovery or adaptation. Curr Opin Cell Biol 2007;19:238–245.
Moynahan ME, Jasin M . Mitotic homologous recombination maintains genomic stability and suppresses tumorigenesis. Nat Rev Mol Cell Biol 2010;11:196–207.
Negrini S, Gorgoulis VG, Halazonetis TD . Genomic instability—an evolving hallmark of cancer. Nat Rev Mol Cell Biol 2010;11:220–228.
Bekker-Jensen S, Mailand N . Assembly and function of DNA double-strand break repair foci in mammalian cells. DNA Repair (Amst) 2010;9:1219–1228.
Huen MS, Chen J . Assembly of checkpoint and repair machineries at DNA damage sites. Trends Biochem Sci 2010;35:101–108.
Huen MS, Grant R, Manke I et al. RNF8 transduces the DNA-damage signal via histone ubiquitylation and checkpoint protein assembly. Cell 2007;131:901–914.
Mailand N, Bekker-Jensen S, Faustrup H et al. RNF8 ubiquitylates histones at DNA double-strand breaks and promotes assembly of repair proteins. Cell 2007;131:887–900.
Kolas NK, Chapman JR, Nakada S et al. Orchestration of the DNA-damage response by the RNF8 ubiquitin ligase. Science 2007;318:1637–1640.
Wang B, Elledge SJ . Ubc13/Rnf8 ubiquitin ligases control foci formation of the Rap80/Abraxas/Brca1/Brcc36 complex in response to DNA damage. Proc Natl Acad Sci USA 2007;104:20759–20763.
Kudoh A, Iwahori S, Sato Y et al. Homologous recombinational repair factors are recruited and loaded onto the viral DNA genome in Epstein–Barr virus replication compartments. J Virol 2009;83:6641–6651.
Kudoh A, Fujita M, Zhang L et al. Epstein–Barr virus lytic replication elicits ATM checkpoint signal transduction while providing an S-phase-like cellular environment. J Biol Chem 2005;280:8156–8163.
Bailey SG, Verrall E, Schelcher C et al. Functional interaction between Epstein–Barr virus replication protein Zta and host DNA damage response protein 53BP1. J Virol 2009;83:11116–11122.
Chen CC, Yang YC, Wang WH et al. Enhancement of Zta-activated lytic transcription of Epstein–Barr virus by Ku80. J Gen Virol 2011;92:661–668.
Dreyfus DH, Liu Y, Ghoda LY et al. Analysis of an ankyrin-like region in Epstein–Barr Virus encoded (EBV) BZLF-1 (ZEBRA) protein: implications for interactions with NF-kappaB and p53. Virol J 2011;8:422.
Sato Y, Shirata N, Kudoh A et al. Expression of Epstein–Barr virus BZLF1 immediate-early protein induces p53 degradation independent of MDM2, leading to repression of p53-mediated transcription. Virology 2009;388:204–211.
Mauser A, Saito S, Appella E et al. The Epstein–Barr virus immediate-early protein BZLF1 regulates p53 function through multiple mechanisms. J Virol 2002;76:12503–12512.
Yao KT, Zhang HY, Zhu HC et al. Establishment and characterization of two epithelial tumor cell lines (HNE-1 and HONE-1) latently infected with Epstein–Barr virus and derived from nasopharyngeal carcinomas. Int J Cancer 1990;45:83–89.
Cheung ST, Huang DP, Hui AB et al. Nasopharyngeal carcinoma cell line (C666-1) consistently harbouring Epstein–Barr virus. Int J Cancer 1999;83:121–126.
Huang DP, Ho JH, Poon YF et al. Establishment of a cell line (NPC/HK1) from a differentiated squamous carcinoma of the nasopharynx. Int J Cancer 1980;26:127–132.
Lo AK, Lo KW, Tsao SW et al. Epstein–Barr virus infection alters cellular signal cascades in human nasopharyngeal epithelial cells. Neoplasia 2006;8:173–180.
Hu Y, Scully R, Sobhian B et al. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev 2011;25:685–700.
Moynahan ME, Pierce AJ, Jasin M . BRCA2 is required for homology-directed repair of chromosomal breaks. Mol Cell 2001;7:263–272.
Galanty Y, Belotserkovskaya R, Coates J et al. RNF4, a SUMO-targeted ubiquitin E3 ligase, promotes DNA double-strand break repair. Genes Dev 2012;26:1179–1195.
Lukas J, Lukas C, Bartek J . More than just a focus: the chromatin response to DNA damage and its role in genome integrity maintenance. Nat Cell Biol 2011;13:1161–1169.
Huen MS, Huang J, Leung JW et al. Regulation of chromatin architecture by the PWWP domain-containing DNA damage-responsive factor EXPAND1/MUM1. Mol Cell 2010;37:854–864.
Lukas C, Savic V, Bekker-Jensen S et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat Cell Biol 2011;13:243–253.
Sonoda E, Hochegger H, Saberi A et al. Differential usage of non-homologous end-joining and homologous recombination in double strand break repair. DNA Repair (Amst) 2006;5:1021–1029.
Pierce AJ, Johnson RD, Thompson LH et al. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev 1999;13:2633–2638.
Bothmer A, Robbiani DF, Di Virgilio M et al. Regulation of DNA end joining, resection, and immunoglobulin class switch recombination by 53BP1. Mol Cell 2011;42:319–329.
Feng L, Chen J . The E3 ligase RNF8 regulates KU80 removal and NHEJ repair. Nat Struct Mol Biol 2012;19:201–206.
Kastan MB, Bartek J . Cell-cycle checkpoints and cancer. Nature 2004;432:316–323.
Bakkenist CJ, Kastan MB . DNA damage activates ATM through intermolecular autophosphorylation and dimer dissociation. Nature 2003;421:499–506.
Kurz EU, Lees-Miller SP . DNA damage-induced activation of ATM and ATM-dependent signaling pathways. DNA Repair (Amst) 2004;3:889–900.
Burma S, Chen BP, Murphy M et al. ATM phosphorylates histone H2AX in response to DNA double-strand breaks. J Biol Chem 2001;276:42462–42467.
Wang B, Matsuoka S, Carpenter PB et al. 53BP1, a mediator of the DNA damage checkpoint. Science 2002;298:1435–1438.
DiTullio RA Jr ., Mochan TA, Venere M et al. 53BP1 functions in an ATM-dependent checkpoint pathway that is constitutively activated in human cancer. Nat Cell Biol 2002;4:998–1002.
Cayrol C, Flemington EK . The Epstein–Barr virus bZIP transcription factor Zta causes G0/G1 cell cycle arrest through induction of cyclin-dependent kinase inhibitors. EMBO J 1996;15:2748–2759.
Mauser A, Holley-Guthrie E, Zanation A et al. The Epstein–Barr virus immediate-early protein BZLF1 induces expression of E2F-1 and other proteins involved in cell cycle progression in primary keratinocytes and gastric carcinoma cells. J Virol 2002;76:12543–12552.
Mauser A, Holley-Guthrie E, Simpson D et al. The Epstein–Barr virus immediate-early protein BZLF1 induces both a G(2) and a mitotic block. J Virol 2002;76:10030–10037.
Hendzel MJ, Wei Y, Mancini MA et al. Mitosis-specific phosphorylation of histone H3 initiates primarily within pericentromeric heterochromatin during G2 and spreads in an ordered fashion coincident with mitotic chromosome condensation. Chromosoma 1997;106:348–360.
Schuler M, Rupa DS, Eastmond DA . A critical evaluation of centromeric labeling to distinguish micronuclei induced by chromosomal loss and breakage in vitro. Mutat Res 1997;392:81–95.
Fenech M . The micronucleus assay determination of chromosomal level DNA damage. Methods Mol Biol 2008;410:185–216.
Difilippantonio S, Gapud E, Wong N et al. 53BP1 facilitates long-range DNA end-joining during V(D)J recombination. Nature 2008;456:529–533.
McFadden K, Luftig MA . Interplay between DNA tumor viruses and the host DNA damage response. Curr Top Microbiol Immunol 2013;371:229–257.
Xie A, Hartlerode A, Stucki M et al. Distinct roles of chromatin-associated proteins MDC1 and 53BP1 in mammalian double-strand break repair. Mol Cell 2007;28:1045–1057.
Dimitrova N, Chen YC, Spector DL et al. 53BP1 promotes non-homologous end joining of telomeres by increasing chromatin mobility. Nature 2008;456:524–528.
Inman GJ, Binne UK, Parker GA et al. Activators of the Epstein–Barr virus lytic program concomitantly induce apoptosis, but lytic gene expression protects from cell death. J Virol 2001;75:2400–2410.
Foghsgaard L, Jaattela M . The ability of BHRF1 to inhibit apoptosis is dependent on stimulus and cell type. J Virol 1997;71:7509–7517.
Matusali G, Arena G, De Leo A et al. Inhibition of p38 MAP kinase pathway induces apoptosis and prevents Epstein–Barr virus reactivation in Raji cells exposed to lytic cycle inducing compounds. Mol Cancer 2009;8:18.
Hui KF, Chiang AK . Suberoylanilide hydroxamic acid induces viral lytic cycle in Epstein–Barr virus-positive epithelial malignancies and mediates enhanced cell death. Int J Cancer 2010;126:2479–2489.
Weitzman MD, Lilley CE, Chaurushiya MS . Genomes in conflict: maintaining genome integrity during virus infection. Annu Rev Microbiol 2010;64:61–81.
Chaurushiya MS, Lilley CE, Aslanian A et al. Viral E3 ubiquitin ligase-mediated degradation of a cellular E3: viral mimicry of a cellular phosphorylation mark targets the RNF8 FHA domain. Mol Cell 2012;46:79–90.
Gaspar M, Shenk T . Human cytomegalovirus inhibits a DNA damage response by mislocalizing checkpoint proteins. Proc Natl Acad Sci USA 2006;103:2821–2826.
Shin YC, Nakamura H, Liang X et al. Inhibition of the ATM/p53 signal transduction pathway by Kaposi's sarcoma-associated herpesvirus interferon regulatory factor 1. J Virol 2006;80:2257–2266.
Carson CT, Orazio NI, Lee DV et al. Mislocalization of the MRN complex prevents ATR signaling during adenovirus infection. EMBO J 2009;28:652–662.
Carson CT, Schwartz RA, Stracker TH et al. The Mre11 complex is required for ATM activation and the G2/M checkpoint. EMBO J 2003;22:6610–6620.
Hagemeier SR, Barlow EA, Meng Q et al. The cellular ataxia telangiectasia-mutated (ATM) kinase promotes Epstein–Barr virus (EBV) lytic reactivation in response to multiple different types of lytic-inducing stimuli. J Virol 2012.
Chen YR, Liu MT, Chang YT et al. Epstein–Barr virus latent membrane protein 1 represses DNA repair through the PI3K/Akt/FOXO3a pathway in human epithelial cells. J Virol 2008;82:8124–8137.
Liu MT, Chen YR, Chen SC et al. Epstein–Barr virus latent membrane protein 1 induces micronucleus formation, represses DNA repair and enhances sensitivity to DNA-damaging agents in human epithelial cells. Oncogene 2004;23:2531–2539.
Kamranvar SA, Masucci MG . The Epstein–Barr virus nuclear antigen-1 promotes telomere dysfunction via induction of oxidative stress. Leukemia 2011;25:1017–1025.
Parker GA, Touitou R, Allday MJ . Epstein–Barr virus EBNA3C can disrupt multiple cell cycle checkpoints and induce nuclear division divorced from cytokinesis. Oncogene 2000;19:700–709.
Chang YH, Lee CP, Su MT et al. Epstein–Barr virus BGLF4 kinase retards cellular S-phase progression and induces chromosomal abnormality. PLoS One 2012;7:e39217.
Acknowledgements
We thank Dr Maria Jasin of Memorial Sloan-Kettering Cancer Center for providing the DR-GFP U2OS cell line and Dr Wolfgang Hammerschmidt of German Research Center for Environmental Health for providing the BZLF1 expression plasmid p509. We also thank the Faculty Core Facility (Faculty of Medicine, University of Hong Kong) for supporting the imaging and flow cytometric studies. This study was supported by the General Research Fund Grant GRF 779312 from Research Grant Council; Area of Excellence on Nasopharyngeal Carcinoma Grant AoE/M-06/08 from Research Grant Council; Committee on Research and Conference grant from the University of Hong Kong; SK Yee Medical Research Fund (to MS-YH); and by Health and Medical Research Fund (12110782).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Laboratory Investigation website
The Epstein-Barr virus (EBV) immediate-early protein BZLF1 is a key mediator in the switch of EBV infection from the latent to the lytic form. In this study, expression of BZLF1 is shown to impair DNA damage response pathways in host cells by blocking the recruitment of 53BP1, a regulator of DNA repair. BZLF1 may function as a link between lytic EBV infection and faulty DNA repair, thus contributing to the carcinogenesis of EBV-associated human epithelial malignancies.
Supplementary information
Rights and permissions
About this article
Cite this article
Yang, J., Deng, W., Hau, P. et al. Epstein–Barr virus BZLF1 protein impairs accumulation of host DNA damage proteins at damage sites in response to DNA damage. Lab Invest 95, 937–950 (2015). https://doi.org/10.1038/labinvest.2015.69
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2015.69
This article is cited by
-
The lytic phase of Epstein–Barr virus plays an important role in tumorigenesis
Virus Genes (2023)
-
Resveratrol inhibits Extranodal NK/T cell lymphoma through activation of DNA damage response pathway
Journal of Experimental & Clinical Cancer Research (2017)
-
Epstein-Barr virus-induced up-regulation of TCAB1 is involved in the DNA damage response in nasopharyngeal carcinoma
Scientific Reports (2017)