Abstract
PAX5 is a nuclear transcription factor required for B cell development, and its expression was evaluated in upper aerodigestive malignancies and pancreatic cancer by immunoblotting. The PAX5 protein expression was relatively strong in small-cell lung cancer (SCLC, 11/12); however, its expression was not detected in non-SCLC (NSCLC, n=13), mesothelioma (n=7), pancreatic (n=6), esophageal (n=6) and head and neck cancer cell lines (n=12). In comparison, PAX8 and PAX3 expressions were absent or non-detectable in SCLC cell lines; however, PAX8 was expressed in most of the tested NSCLC cell lines (13/13) and also frequently in all the other cell lines. We also detected frequent expressions of PAX2 and PAX9 protein in the various cell lines. Utilizing neuroendocrine tumor samples, we found that the frequency as well as the average intensity of the expression of PAX5 increased from pulmonary carcinoid (9%, moderate and strong PAX5 expression, n=44), to large-cell neuroendocrine carcinoma (LCNC, 27%, n=11) to SCLC (33%, n=76). FISH analysis revealed no translocations of the PAX5 gene, but polyploidy in some SCLC tumor tissues (6/37). We determined that PAX5 could regulate the transcription of c-Met using luciferase-coupled reporter and chromatin immunoprecipitation analysis. In addition, the phospho-c-Met (active form) and PAX5 were both localized to the same intra-nuclear compartment in hepatocyte growth factor treated SCLC cells and interacted with each other. Finally, we determined the therapeutic translational potential of PAX5 using PAX5 knockdown SCLC cells in conjunction with Topoisomerase 1 (SN38) and c-Met (SU11274) inhibitors. Loss of endogenous PAX5 significantly decreased the viability of SCLC cells, especially when combined with SN38 or SU11274, and maximum effect was seen when both inhibitors were used. Therefore, we propose that PAX5 could be an important regulator of c-Met transcription and a potential target for therapy in SCLC.
Similar content being viewed by others
Main
It is estimated that there will be over 214 000 patients diagnosed with lung cancer in 2008, and over 160 000 patients will die of this disease in the United States.1 Lung cancer can usually be divided into small-cell lung cancer (SCLC) and non-SCLC (NSCLC) with histological subtypes including adenocarcinoma, bronchioloalveolar carcinoma, squamous cell carcinoma and large-cell carcinoma. The overall 5-year survival of NSCLC, which accounts for 85% of all lung cancers, is 16%. Moreover, SCLC comprises 14% of all lung cancers and the prognosis is only 6% over 5 years. Even with the most current therapeutic agents, the response is relatively poor in a majority of the patients.1 In order to further identify potential therapeutic targets in lung cancer, we examined the expression profile and the role of PAX genes, in particular PAX5 (also known as B cell-specific activator protein) in lung cancer.
PAX (paired box) genes are a family of nine nuclear transcription factors that play a crucial and indispensable role, in various developmental programs, both in vertebrates and invertebrates. All the PAX genes have the characteristic paired domain that is essential for specific DNA binding and, in addition, some have either an octapeptide region or a homeodomain or both. The later two appear to be essential for protein–protein interactions. In humans, all the nine PAX genes are expressed during various stages of embryogenesis and development. In adults, most of the PAX genes are silent; however, they become selectively active during tissue repair and regeneration. Interestingly, several of the PAX genes have been reported to be expressed in various cancers and are likely to contribute to the overall tumorigenesis. Usually, expressions of the PAX genes in cancers appear to be related to tissue lineage thereby suggesting a process of dedifferentiation.2 In this study, we have examined the expression of PAX5 protein and compared it with the expression of other PAX proteins such as PAX 2, 8 and 9.
PAX5 is expressed normally in the developing brain at the boundary of the mid and hind brains and neural tubes. It is essential for B cell development; and its expression has been noted at all stages of B cell development except in the terminally differentiated plasma cells. PAX5 knockout mice predictably lack B cells and therefore any humoral immunity.3 In addition, they also have defective inferior colliculus and anterior cerebellum. Significant PAX5 expression has been noted in most of the B cell lymphomas (B cell chronic lymphocytic leukemia, mantle cell leukemia and follicular lymphoma); however, the T and null-cell lymphomas, as well as plasmacytomas and multiple myeloma lack the PAX5 expression.4 The deregulated expression of PAX5 has also been noted in pediatric cancers such as medulloblastomas, and its expression in normal cells is correlated inversely with neuronal differentiation.5 Most importantly, significant PAX5 expression has been noted in tumors of neuroendocrine origin such as neuroblastoma and SCLC.6 PAX5 was found to be over expressed in aggressive neuroblastoma (N-type) as opposed to the less aggressive S-type. A similar scenario has been reported with respect to highly metastatic SCLC cell lines. Significant amounts of PAX5 transcripts were found to be present in several SCLC cell lines but not in NSCLC cell lines.6 As the enforced expression of PAX5 in neuroblastoma S-type cells confers on them a more oncogenic phenotype, and because the PAX5 knockdown results in a significant loss in cell viability, PAX5 is believed to not only support cancer cell survival but also contribute to metastasis.
Mechanisms underlying metastasis are extremely complex and the contributing factors could be many. The receptor tyrosine kinase (RTK) c-Met is known to play a significant role in cell motility and tumor metastasis.7 We and others have shown that c-Met is over expressed in SCLC and plays a significant role in lung tumorigenesis and metastasis.8, 9, 10 PAX3 is known to regulate transcription of c-Met in limb muscle in which both are expressed. PAX3 and its oncogenic fusion product PAX3-FKHR are both known to positively regulate the transcription of c-Met.11 It is not known whether PAX5 expressed in SCLC also enhance c-Met transcription. Therefore, we examined the PAX5 protein expression in SCLC tumor tissues and its role in the regulation of c-Met expression.
In this study, using cell line panels that represent SCLC and other cancers, we show that the PAX5 protein expression is restricted to SCLC cell lines and its expression is not detectable in NSCLC, pancreatic, mesothelioma, head and neck and esophageal cancer cell lines. In comparison, PAX8 protein was expressed in NSCLC but not SCLC cell lines; however, its expression was seen with varying frequency in other cancer cell lines. We also show here that the PAX5 protein expression increased from carcinoid to large cell to SCLC tumors, suggesting a positive correlation between intensity and frequency of the PAX5 expression and metastatic potential. In addition, we show that PAX5 is a direct transcriptional activator of the RTK c-Met. The results presented here show PAX5 to be associated temporally and spatially with c-Met and thus, it can serve as a potential therapeutic target in SCLC.
MATERIALS AND METHODS
Antibodies and Other Reagents
PAX 2, 5, 8 and 9 polyclonal antibodies were from Abcam (Cambridge, MA) and PAX5 monoclonal antibody was from BD Biosciences (Franklin Lakes, NJ). The phospho-specific (pY1230/1234/1235) anti-c-Met rabbit polyclonal was from Biosource (Carlsbad, CA), and the rabbit polyclonal against Ron-β and the mouse monoclonal against Topoisomerase 1 were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). SU11274 (3Z)-N-(3-Chlorophenyl)-3-((3,5-dimethyl-4-((4-methylpiperazin-1-yl)carbonyl)-1H-pyrrol-2-yl)methylene)-N-methyl-2-oxo-2,3-dihydro-1H-indole-5-sulfonamide, the c-Met small molecule inhibitor was from EMD Calbiochem (San Diego, CA) and SN38, the active metabolite of the topoisomerase 1 inhibitor irinotecan was purchased from the American Custom Chemicals corporation (San Diego, CA). A set of four different small interfering RNAs (siRNAs) specific for PAX5 and the control RNA were purchased from Qiagen (Cambridge, MA).
Cell Lines
The SCLC, NSCLC, mesothelioma, esophageal, pancreatic and head and neck cancer cell lines were obtained from the American Type Culture Collection (Manassas, VA) and were cultured in RPMI 1640 medium (Gibco/BRL) supplemented with 10% (v/v) fetal bovine serum supplemented with L-glutamine and 1%(v/v)penicillin–streptomycin at 37°C with 5% CO2.
Cell lysis and Immunoblotting
Whole cell lysates were prepared in RIPA lysis buffer (50 mM Tris (pH 8.0), 150 mM Nacl, 10%glycerol, 1% NP-40, 0.5% Sodium deoxycholate, 0.1% SDS and 0.42% NaF) containing protease inhibitors (1 mM phenylmethylsulfonyl fluoride, 1 mM Na3VO4, 5 μg/ml leupeptin).12 Protein concentrations were determined using the Bradford Assay, and 60–80 μg protein lysates were separated by 10% SDS-PAGE under reducing conditions and transferred to PVDF membranes (Millipore, Bedford, MA). The membranes were blocked in 5% non-fat dried milk, freshly made in TBST. Proteins were detected by immunoblotting using an enhanced chemiluminescense reagent (Boston Bioproducts, Worcester, MA).12
Immunoprecipitation Experiment
H249 and H69 (SCLC) cells were lysed in a mild detergent, and immunoprecipitations with anti-c-Met, anti-PAX5 and normal mouse IgG were carried out as described earlier.12 The immune complexes were then collected and the proteins separated and probed by immunoblotting as described above.
Plasmids and Vectors
MET promoter linked to luciferase reporter
A fragment of the MET promoter (−535 to −172 upstream of the transcriptional start site) was amplified from normal human genomic DNA and subcloned into pGL2-basic vector (Promega, Madison, WI) to create the reporter plasmid wt hMET pm. The MET1 (see Figure 5a) consensus site for paired protein binding was mutated by site-directed mutagenesis, replacing the sequence GTCCCGC to ACTAGTC, thereby destroying the MET1 site and creating a SpeI restriction site (hMET pm Δ). The human PAX5 expression construct was kindly provided by Dr Meinrad Busslinger (Research Institute of Molecular Pathology, Austria). HEK 293T cells were transfected with either wt hMET pm or hMET pm, Δ reporter constructs, an internal control β-galactosidase expressing construct pCMV (Clontech, Mountain View, CA) and the presence or absence of PAX5 expressing construct. The total amount of DNA transfected was kept constant by the addition of pBluescript plasmid (Stratagene, Cedar Creek, TX). Cells were transfected according to the manufacturer's protocols (Effectene reagent, Qiagen). After 48 h post-transfection, luciferase and β-galactosidase assays (Promega) were performed. Luciferase activity was normalized to β-galactosidase activity levels as an internal control. All experiments were performed in triplicate.13
Chromatin Immunoprecipitation (ChIP) Assay
H69 cell suspensions were fixed in 1% formaldehyde and quenched in 0.125 M glycine, then processed according to the manufacturer's protocol (Upstate Biotechnology, Lake Placid, NY). For immunoprecipitation of PAX5–DNA complexes, 1 μl of mouse monoclonal antibody (BD Biosciences) was added per experimental reaction. A non-specific antibody, normal IgG (Sigma), was used as a negative control against non-specific DNA precipitation by an antibody.13 Nested PCR was performed with primers to the Met enhancer, first with TCCGCCTCTAACAATGAACTCC (F) and AAGGTGAAACTTTCTAGGTGG (R) then a second PCR reaction with primers TGCCCAAATCTCTCTAAACCC (F) and AAGTTTTCTCGCCCTGGCTGCG. All chromatin immunoprecipitation (ChIP) samples were tested for false-positive PCR amplification using primers that amplify sequence from within the fourth large coding exon of the β-tubulin gene to control against genomic DNA contamination. The nested primer set for this control was AAAGGCCACTACACAGAGGG (F) and TACCAACTGATGGACGGAGAGG (R) for the first PCR reaction, then TTGATTCTGTCCTGGATGTGG (F) and TCAGACACTTTGGGTGAAGGC(R) for the second PCR round. For the first PCR reaction, 10% of the ChIP sample was utilized as template. For the nested second reaction, 5% of the first PCR reaction was used as template.
Transfection with Small Interfering RNA (siRNA)
Four different siRNAs targeting human PAX5 mRNA were obtained from Qiagen and pooled the PAX5 expression to silence. For each experiment, 4 × 105 cells/well were plated in 6-well plates and transfected with 100 nM siRNA (cocktail) or scrambled control RNA (scRNA) for 96 h using HiPerfect Transfection Reagent according to the manufacturer's instructions (Qiagen). After 96 h incubation with siRNA, the cell lysates were prepared using RIPA lysis buffer.
Immunofluorescence and Confocal Microscopy
Cells were grown in a medium devoid of FBS on glass cover slips (coated with polylysine) in 6-well plates overnight. They were treated with hepatocyte growth factor (HGF) (40 ng/ml) for various times and fixed in paraformaldehyde (3.7%) in PBS for 15 min at 37°C, and then permeabilized with Triton X-100 in PBS (0.1%) for 10 min at room temperature. Fixed cells were washed twice with ice-cold PBS and once with PBS containing Triton X-100. They were then incubated with primary antibodies in 1% BSA for 1 h followed by three washes with PBS. Further incubation with the appropriate secondary antibodies conjugated with fluorescein (CY3 and CY5) followed. After washes, the samples were mounted in Vectashield.
DNA Extraction from Paraffin-Embedded Tissue
Three 10 μM-thick sections were cut from each paraffin-embedded tissue block and placed into a microcentrifuge using clean forceps. The microtome blade, tweezers and other equipments that could come into contact with the sample were sterilized carefully before processing each tissue block. Care was taken to minimize cross contamination. DNA extraction was carried out according to Sato et al, with minor modifications.14
Tissue Microarrays (TMAs)
TMAs were assembled at the University of Chicago, Chicago, IL and The Methodist Hospital, Houston, TX. For the TMA, 44 carcinoids, 76 SCLC and 11 large-cell neuroendocrine carcinoma (LCNC) were identified from tissue archives. A minimum of three tissue cores with a diameter of 1 mm was arrayed into a recipient block using an automated tissue microarrayer ATA-27 (Beecher Instruments, Sun Prairie, WI). All tissues were obtained under protocols approved by applicable IRBs and they were used for PAX5 staining.
TMA paraffin sections were deparaffinized in xylene, rehydrated through graded ethanol solutions to distilled water and then washed in Tris-buffered saline. Antigen retrieval was carried out by heating sections in ETDA buffer (pH=9) for 15 min in a microwave oven. Endogenous peroxidase activity was quenched by incubation in 3% H2O2 in methanol for 5 min. Non-specific binding sites were blocked using Protein Block (DAKO, Carpinteria, CA) for 20 min. Then the tissue sections were incubated for 1 h at room temperature with the mouse monoclonal antibody against PAX5 (clone 24, 1:25 dilution, BD Biosciences). This step was followed by 30 min incubation with goat anti-mouse IgG conjugated to a horseradish peroxidase-labeled polymer (Envision™+ System, DakoCytomation, Carpinteria, CA). Slides were then developed for 5 min with 3-3′-diaminobenzidine chromogen, counterstained with hematoxylin. Negative controls were performed by substituting the primary antibody step with non-immune mouse immunoglobulins. Scoring of the immunostaining intensity in the nuclei was performed as follows: 0, no staining; 1+, weak staining; 2+, moderate staining; and 3+, strong staining (as high as seen in normal B cells used as internal positive control).
FISH Analysis
FISH on paraffin-embedded tissue sections was performed, as described earlier,15, 16 using probes that were constructed by random priming (Bioprime, Invitrogen, Carlsbad, CA) of purified DNA from bacterial artificial chromosomes RP11-1009K17, RP11-844C13, RP11-591B19, RP11-12P15, RP11-614P24, RP11-397D12, RP11-660F1 and RP11-643F16 that flank the PAX5 gene. Briefly, bacterial artificial chromosomes with DNA inserts immediately upstream and downstream of the PAX5 gene were differentially labeled with dCTP-biotin (Bioprime) or dUTP-digoxigenin (Roche, Nutley, NJ), hybridized against small-cell carcinoma tissues that were digested with Digest-All 3 (Zymed) and reacted with Streptavidin-Alexa594 (Molecular Probes, Carlsbad, CA) and anti-digoxigenin-fluorescein (Roche) in the presence of CAS block (Zymed, San Francisco, CA) for detection. All bacterial artificial chromosome probes hybridized to the PAX5 locus at 9p13 on metaphase spreads from normal human lymphocytes (unpublished data). Hybridizations were visualized on a Ziess Axioplan2 ie, microscope with Zeiss Neofluar and ApoFluar objectives and a megapixel CCD camera using Metasystems software (Metasystems, GmbH, Altlussheim, Germany).
Sequencing and Mutation Analysis of PAX5
The individual exons of PAX5 in genomic DNA were amplified by Multiplex PCR reagent (Qiagen, Hilden, Germany). In each multiplex, PCR reaction up to four sets of amplification primers were used. For DNA isolated from the paraffin-embedded tissue, the amplicon size was kept at <500 bp. PCR was performed in 15 μl volumes containing 1 × buffer, 200 nM of each primer and 100–200 ng of template DNA. Genomic DNA obtained from cell line or of archival formalin-fixed, paraffin-embedded tumor tissue was used as template. Primer cocktails were prepared from 50 μM stock solutions. PCR profile: 95°C for 15 min, then 35 cycles of 94°C for 30 s, 60°C for 1 min and 72°C for 1 min, followed by one cycle of 72°C for 10 min. PCR products were treated with ExoSAP-IT (USB Corp., Cleveland, OH) and sequenced employing Big Dye Terminator Chemistry (Applied Biosystems, Weiterstadt, Germany). Sequencing was performed on the forward coding strand. Confirmation of nucleotide alterations was performed by repeating PCR followed by sequencing the forward and reverse strands. Chromatograms were analyzed for mutations using Mutation Surveyor v2.61 (Softgenetics, State College, PA). The nucleotide positions numbering is relative to the first base of the translational initiation codon according to the full-length PAX5 cDNA (CCDS 6607.1).
PAX5 PCR-Amplification Primers

Cell Viability
In order to evaluate the inhibition of PAX5-affected cell growth, 4 × 104 H69 (SCLC) cells were plated in 6-well plates in duplicates, and grown in serum-containing media for 24 h. siRNA against human PAX5 (as above) was used at a final concentration of 100 nM. Transfections were achieved utilizing Fugene HD (Roche Diagnostics, Basel, Switzerland), as per the manufacturer's instructions. SU11274 was used to inhibit c-Met at 2.5 μM, whereas SN38 was used at 2.5 nM. All treatments were performed for 72 h in serum-free media. Cell growth was estimated utilizing fluorometric readings after the addition of Alamar Blue, a non-radioactive, non-toxic dye that is reduced. This reduced compound emits fluorescence that is proportional to the metabolic activity. A HT Synergy Plus microplate reader (Biotek, Winooski, VT) was used to measure fluorescence. Readings were normalized to control wells.
Statistics
Statistical analysis was performed utilizing Graphpad Prism version 5.0 (Graphpad Inc, San Diego, CA). In order to evaluate for statistically significant differences between two continuous variables the unpaired Student's t-test was used. Differences were considered significant when ‘P’ values were <0.05.
RESULTS
Selective Expression of PAX5 in SCLC
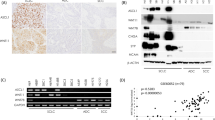
Using panels of cancer cell lines that represent lung (SCLC, n=12 and NSCLC, n=13), head and neck (n=12), mesothelioma (n=7), esophageal (n=6) and pancreatic cancers (n=6), we determined the relative protein expression levels of PAX5 and compared it with other PAX protein levels using whole cell lysates and standard immunoblotting techniques. The level of actin expression in the same sample served as loading control. Immunoblots of various PAX proteins were performed (Figure 1), the relative protein expression levels were scored visually and presented in the Table 1 to facilitate comparisons.
Expression profile of PAX5 in SCLC, NSCLC, mesothelioma, esophagus, pancreas and head and neck cancers. Each panel represents a collection of cancer cell lines that signify a particular cancer. The whole cell lysates were immunoblotted with anti-PAX5, anti-PAX2, anti-PAX8 and anti-PAX9 antibodies. The same sample set was also used in parallel to determine the c-Met, RON and Topo1 expressions using specific antibodies in immunoblotting. β-actin level served as loading control.
PAX5 protein was expressed in most of the SCLC cancer lines (n=11/12) and the levels were non-detectable in all other cancer cell lines. Half of the SCLC cell lines examined expressed relatively high levels of PAX5. SCLC cell lines also expressed varying amounts of PAX2 protein and only 5/12 SCLC cell lines expressed PAX9. Interestingly, none of them expressed PAX3, or PAX7 (data not shown). In comparison, all the NSCLC cell lines (n=13) expressed PAX8. The majority of the NSCLC cell lines also expressed PAX2; however, the PAX9 expression was seen only in five NSCLC cell lines.
In pancreatic, head and neck, esophageal cancers and mesotheliomas, no particular PAX protein was expressed exclusively in any of these cancers. Most of them were positive for one or more of PAX proteins, excluding PAX5 (with validation from internal controls such as Raji cells, data not shown). These tumor cell lines expressed varying combinations of PAX2, 8 and 9 proteins. In the seven mesothelioma and six pancreatic cancer cell lines examined, the above three PAX proteins were expressed in all but one. We also failed to detect any PAX8 protein in all the six esophageal cancer cell lines. An important finding was the expression of PAX9 in all the pancreatic cancer cell lines examined and its more frequent appearance in other cancers.
In the same samples, we also determined the relative protein expression levels of potential therapeutic targets such as the RTKs, c-Met (and a related kinase RON) that is known to be regulated by PAX3 and the DNA topology enzyme Topoisomerase 1 (Topo1). The majority of the cancer cell lines that represented the six different cancers expressed c-Met; however, RON was less frequently seen in mesothelioma ((2/7) and pancreatic cancer (3/6) cell lines. Topo1 was found to be expressed in most of the cancer cell lines that represented SCLC (11/12), NSCLC (12/13), pancreas (5/6), esophageal (5/6) and head and neck cancer cell lines (10/12); however, significant Topo1 expression was observed only in three out of the six mesothelioma cell lines.
Frequency and Intensity of PAX5 Expression Increases with Metastatic Potential of Neuroendocrine Lung Tumors
In order to validate the expression of PAX5 in tumors of neuroendocrine origin, we determined the protein expression in various archival lung tumors (with tumor TMAs). Utilizing anti-PAX5 antibody in immunohistochemistry, we determined the PAX5 expression in pulmonary carcinoid (n=44), large-cell neuroendocrine cancer (LCNC) (n=11) and SCLC (n=76). A representative staining is shown in Figure 2a. The results obtained from a total of 131 lung cancer tumor samples are summarized in Figure 2b–e. The percentage of PAX5-positive tumor samples increased from 27% in carcinoids to 55% in LCNC and 66% in SCLC. Based on the intensity of the expression of PAX5 in the nucleus, we scored each tumor sample as low (+1), medium (+2) and maximum (+3). The percentage of positive cells in each of the three above categories also increased significantly from carcinoid to LCNC and to SCLC (chi-square, P=0.007), and overall reflects increased metastatic potential.
Expression of PAX5 in lung tumor tissues of neuroendocrine origin. (a) Nuclear PAX5 staining was carried out as described in Materials and Methods. Immunohistochemistry images represent negative (0), weak (1+), moderate (2+) and strongly positive (3+) PAX5 expression of different tumor subtypes. (b–d) Pie charts show relative proportion of negative (blue), weak-positive (yellow), moderate-positive (red) and strong-positive cases (maroon) in carcinoid, LCNC and SCLC tumor samples. (e) PAX5 expression levels and relative frequency based on independent scoring by two pathologists. It was lowest in lung carcinoids (top panel, 73% negative cases, n=44), intermediate in large-cell neuroendocrine carcinomas (middle panel, 45% negative cases, n=11) and highest in small-cell carcinomas (bottom panel, 34% negative cases, n=76). A chi-square test shows statistically significant increased PAX5 expression from carcinoid to LCNC to SCLC (P=0.007).
PAX5 Gene Copy Number in SCLC
We ascertained by interphase fluorescence in situ hybridization (FISH) the presence of rearrangements and copy number changes in the PAX5 gene to determine the mechanism of PAX5 over expression in SCLC. The paraffin-embedded tissue sections of SCLC were hybridized to dual-color FISH probes that were constructed from bacterial artificial chromosomes that flank PAX5 on chromosome 9q13 (Figure 3a and b). No split FISH signals that are indicative of PAX5 rearrangements were detected in SCLC from 30 different patients (Figure 3c and d). On the other hand, changes in PAX5 copy number were present in 21 (70%) of the SCLC. Fifteen (50%) SCLC exhibited increased PAX5 copy number (N3, N4 and/or N5), whereas one (3%) additional small-cell carcinoma contained PAX5 tetraploid (N4) and octaploid (N8) cells. In contrast, five (17%) SCLC exhibited decreased PAX5 copy number (combined N1 and N2) and nine (30%) SCLCs were normal diploid at the PAX5 locus (N2). No correlation between PAX5 copy number and clinical-pathologic parameters was observed in these patients (data not shown).
PAX5 locus on human chromosome 9 and FISH analysis. (a) The PAX5 gene, located on chromosome 9 on p13, and involved in t(9;14)(p13;q32) translocations recurring in small lymphocytic lymphomas of the plasmacytoid subtype. (b) The PAX5 gene locus and the digitonin and biotin-labeled BAC constructs used in the FISH are represented schematically. (c) Flowchart shows SCLC tissue sample processing for FISH analysis. (d) Representative micrograph of a SCLC tissue FISH analysis.
PAX5 Mutational Analysis in SCLC
All 10 exons coding for the full-length PAX5 were sequenced using aqueous extracts of genomic DNA isolated from 21 SCLC cell lines and 24 SCLC tumor tissue specimens, as described in Materials and Methods. As shown in Figure 4, the frequency of occurrence of either synonymous or non-synonymous mutations was not significant. One non-synonymous heterozygous mutation 964G>AG leading to amino acid change A322S in the PAX5 gene was detected in cell line H146. Also, another heterozygous synonymous mutation 1029G>AG corresponding to amino acid G343 was detected in two specimens out of 24. These mutations were either in the transactivation domain or within its proximity. In the tested SCLC tissue samples, non-synonymous mutations were not detected.
PAX5 gene mutational analysis. (a) PAX5 functional domains such as PD, DNA binding paired domain; OP, conserved octapeptide; HD, partial homeodomain; TA, transactivation domain; ID, inhibitory domain are schematically represented. (b) Exons that code various domains in PAX5 protein are shown. (c) Summary of mutations detected in SCLC cell lines and tissues. The PAX5 protein coding exons were PCR amplified and sequenced using genomic DNA from 21 SCLC cell lines and 24 SCLC tumor tissues. A non-synonymous heterozygous mutation 964G>AG resulting in an amino acid change A322S of the TA domain (amino acids 304–358) was detected in cell line H146. All the other mutations were synonymous.
PAX5 Directly Promotes Transcription of the RTK, c-Met
As SCLC is the most aggressive of all cancers, and also because these tumors express relatively high levels of c-Met, a RTK that is known to play a significant role in both tumor growth and metastasis,1, 7 we investigated the relationship between the nuclear transcription factor PAX5 and c-Met. In addition, a highly related PAX3 is known to directly activate transcription of c-Met promoter.11 The paired binding site defined for PAX3 in this enhancer, GTCCCGCTT, is also similar to sites described for PAX5, and the details are shown in Figure 5a. In order to test whether PAX5 can enhance the luciferase reporter linked to c-Met promoter, we generated a mammalian expression plasmid that contained hMET promoter linked to luciferase and also a similar construct, in which the putative PAX5 binding site was deleted in the c-Met promoter sequence. We tested the ability of PAX5 to activate the expression of reporter gene in both reporter constructs by transfecting HEK 293T cells with or without expression plasmids encoding PAX5 and measuring luciferase activity. Compared with control-transfected cells, PAX5 induced significant increases in luciferase activity from the wt hMET promoter construct (6.39±1.99 light units). Mutating the PAX5 binding site completely abolished the ability of PAX5 to activate reporter activity (Figure 5b). To determine whether PAX5 associates with the MET enhancer in small-cell lung carcinoma cells, we performed ChIP assays. DNA–protein complexes within H69 cells were cross-linked and precipitated with either an antibody specific for PAX5 or normal mouse IgG (Figure 5c). After precipitation and DNA purification, the PCR product was generated using MET promoter primers from samples from the PAX5 antibody ChIP or input, but not with non-specific antibody or with β-tubulin control primers (Figure 5c).
PAX5 is a direct activator of c-Met transcription. (a) PAX5 responsive element in the MET receptor gene. Sequence of the MET1 paired consensus site in the MET promoter and in the reporter construct wt hMET pm is shown. The sequence below has the MET1 site destroyed, GTCCCGC to ACTAGTC. (b) Transfection of HEK 293T cells with both the hMET pm (wild-type MET promoter sequence) and hMET pm D (MET promoter with MET1 site mutated) reporter constructs were carried out and all transfections were normalized for transfection efficiency and the reporter activities were expressed as fold activation compared with transfection without PAX5. (mean±s.d., n=12 for each set). (c) Chromatin immunoprecipitation (ChIP) analysis was performed in H69 SCLC cells. The top panel represents PCR product obtained using primers for the MET promoter enhancer and the bottom panel represents PCR product using primers located in exon 4 of the β-tubulin gene (negative control). Lane 1, precipitation with mouse anti-PAX5 antibody; Lane 2, precipitation with normal mouse IgG; Lane 3, cell lysate input, pre-ChIP; and Lane 4, water blank PCR negative control are shown. (d) H526 SCLC were transfected with scrambled and PAX5-specific siRNA (four different siRNAs pooled) at 100 nM. As shown, loss of PAX5 protein expression in siRNA-treated cells is accompanied by a similar loss in c-Met protein levels.
To show the endogenous relationship between PAX5 activity and c-Met protein levels, we knocked down PAX5 in H526 SCLC cells and determined the intracellular levels of c-Met. As shown in Figure 5d, cells transduced with siRNA that specifically targets PAX5 resulted in a dramatic drop in endogenous PAX5 levels when compared with cells transduced with the control-scrambled siRNA. In addition, loss in the PAX5 protein expression was accompanied by a concomitant loss in the c-Met expression, thereby further substantiating that PAX5 is a direct and positive regulator of c-Met transcription.
PAX5 and c-Met Interact with Each Other in the Nuclear Compartment
Although c-Met is a RTK that is mainly localized to the plasma membrane, we and others have observed partial localization of phospho-c-Met, especially after activation with HGF, to the nucleus in SCLC cells 17, 18, 19 (Kanteti and Salgia, unpublished). As PAX5 is mainly nuclear, we therefore examined the possibility that there could be significant co-localization of c-Met in HGF-treated SCLC cells. Representative confocal pictures showing intracellular location of p-c-Met, using immunofluorescence staining as described in Materials and Methods are shown in Figure 6a. In resting H82 cells, all of the PAX5 expression, as expected, was diffuse and restricted to the nucleus. There was virtually no phospho-c-Met in the cell (left panel). Upon treatment of cells with HGF for 10 min, the nuclear PAX5 staining became more intense and punctuated. At the same time, we also observed a dramatic appearance of phospho-c-Met in the nucleus, and majority of it was found to be co-localized with PAX5 (right panel); thus, c-Met and PAX5 are both linked spatially and temporally.
PAX5 interacts with activated c-Met in SCLC cells upon HGF treatment. (a) Representative confocal pictures showing individual staining patterns for phospho-c-Met and PAX5 and the two merged in untreated (left quadrant) and HGF treated (right quadrant) in H82 cells are shown. The cells were treated for 10 min with HGF (40 ng/ml). (b) Co-immunoprecipitation of phospho-c-Met with PAX5. H249 cells were treated with and without HGF (40 ng/ml) for 10 min and cell lysates prepared by standard method. Immunoprecipitation (200 μg lysates) was carried out using anti-PAX5 antibody that revealed significant amounts of phospho-c-Met only in HGF-treated sample. (c) Immunoprecipitation of c-Met coprecipitated significant amounts of PAX5.
This raised the possibility that PAX5 could interact with c-Met and we verified the same using an independent cell line H249 and H69 (SCLC). As shown in Figure 6b, immunoprecipitation of PAX5 co-precipitated significant amounts of phospho-c-Met, with HGF stimulation and the reverse experiment carried out with H69 cell lysates confirmed the same (Figure 6c).
Loss of PAX5 Significantly Enhances SCLC Cell Death when Combined with Topo1 and c-Met Inhibitors
In order to test the translational potential of PAX5 as an adjuvant chemotherapeutic target in SCLC therapy, as an initial step, we determined the effects of knockdown of PAX5 in H69 cells, individually and in combination with SU11274 (a specific c-Met inhibitor currently in clinical trials) and SN38 (a potent Topoisomerase 1 inhibitor and the active metabolite of irinotecan); a drug used in the treatment of SCLC. The cells were transfected with PAX5 siRNA and treated with SU11274, SN38 or combinations for 3 days, (Figure 7). Loss of endogenous PAX5 in these cells had a marginal adverse effect on the viability of the SCLC cells. The cell viability in the above knockdown cells further decreased when treated with either SU11274 or SN38; however, the loss in viability was significantly higher in those cells treated with the Topo1 inhibitor SN38. Finally, treatment of PAX5 knockdown cells with both SN38 and SU11274 compounds resulted in maximum loss in viability.
Combinatorial effect of PAX5 knock down with c-Met and Topo1 inhibitors in SCLC cells. H69 SCLC cells were transfected with either control or PAX5-specific siRNA duplexes (Qiagen). As shown, they were then treated with either SU11274, or SN38, or a combination of both. The histograms represent percentage of cell viability along with statistical significance.
DISCUSSION
The PAX genes are highly conserved throughout evolution and they play a complex and vital role in the development as well as the repair and regeneration of specific tissues. As cancers mimic, albeit in a chaotic fashion, the process of dedifferentiation, it is therefore not surprising that PAX genes are expressed in various cancers.2 In this study, we have focused mainly on the expression and functional consequence of PAX5 in tumors of neuroendocrine origin such as SCLC. We have shown clearly the preferential expression of PAX5 in SCLC but not in NSCLC, mesothelioma, esophageal, pancreatic and head and neck cancer cell lines (Figure 1 and Table). Further, the PAX5 protein expression data in lung tumors (Figure 2) supported the above observation; and the frequency and intensity of the PAX5 protein expression positively correlated with neuroendocrine tumors. SCLCs are known to be highly metastatic and over express several RTKs such as c-Kit and c-Met. We and others have shown that c-Met plays an important role in both lung tumorigenesis and metastasis.20 A striking observation, we made in this paper, was the demonstration of a temporal and spatial link between the expressions of PAX5 and c-Met proteins in the HGF-treated SCLC cells. In addition, we also showed that PAX5 directly promotes transcription of c-Met. Finally, we also found that loss of the PAX5 expression in SCLC cells synergized with the ability of c-Met and Topo1 inhibitors to kill SCLC cells.
The PAX5 gene plays an indispensable role in B cell development and is expressed at almost all stages except in the terminally differentiated plasma cells.3 Most of the Hodgkin and non-Hodgkin B cell lymphomas express PAX5.21 However, several cancers such as plasma cell neoplasms, multiple myeloma, plasmablastic lymphoma and T cell lymphomas, appear to be negative for the PAX5 expression.22 In addition to B cells, PAX5 is expressed naturally in some parts of the developing nervous system. In this study, we have clearly shown that the PAX5 protein expression is restricted to lung tumors of neuroendocrine origin such as carcinoid, LCNC and SCLC but not general NSCLC. The expression of considerable amounts of PAX5 transcripts in SCLC but not NSCLC cell lines was reported previously by Baumann Kubetzko et al; however, they carried out RT-PCR using RNA preparations from both SCLC and NSCLC cell lines (five from each group).6 Here, we have not only determined the relative levels of the PAX5 protein expression in SCLC cell lines, but we also extended the findings to SCLC tumor tissues and reinforced the idea that the PAX5 expression is seen predominantly in tumors of neuroendocrine origin. Similar observations were made recently by others by carrying out immunohistochemical analysis of PAX5 in needle biopsies of tumors of neuroendocrine origin, and through high-throughput TMA analysis of FFPE tumor sections.23, 24 Using archival paraffin-embedded lung tumor tissues, we were the first to positively correlate the intensity and frequency of the PAX5 expression to their metastatic potential (Figure 2).
In addition to PAX5, we also determined the expressions of PAX2, 8 and 9 in lung cancer and other cancer cell lines. As shown in Figure 1, PAX2 and PAX9 expressions, although not consistent, were seen in most of the cancers examined. To a lesser extent, similar findings were made with respect to PAX8. One interesting fact was that the PAX8 expression was seen in all of the NSCLC cell lines tested but not in any of the SCLC cell lines. Therefore, we propose determination of differential expressions of PAX5 and PAX8 proteins in lung cancers to further distinguish SCLC from NSCLC. Despite the fact that PAX2 and also PAX8 have a nephritic lineage and are over expressed in kidney cancers,25, 26, 27, 28 their expressions were seen in all of the cancers we examined. A similar observation was also made by others (reviewed in 2). In pediatric thyroid cancers, PAX8 is associated with a high degree of recurrence and poor prognosis, and is expressed in differentiated but not undifferentiated cancers;29, 30 however, we showed here that it was expressed in NSCLC, head and neck, pancreatic and esophageal cancers. Here, of all the cancers we examined, SCLC is the most metastatic and all the 12 cell lines from this group failed to express PAX8. Therefore, it is very likely that in addition to PAX8, there could be one or more factors that in combination determine the prognosis. We also determined the relative protein expression levels of PAX3 and PAX7, two genes known to be over expressed and frequently translocated and fused to the fork head receptor protein transcription factor (FKHR) in pediatric alveolar rhabdomyosarcoma;31, 32, 33, 34 however, we failed to detect any PAX3 and PAX7 protein expressions (data not shown).
Here, of all the four PAX proteins examined, the PAX9 expression was the most surprising. Its expression was first reported in esophageal cancers. Progressive loss of the PAX9 expression was found to correlate with increasing malignancy and dysplasia of cancerous esophageal epithelium.35 In general, squamous cell cancers of the esophagus appear to be positive for PAX9. In comparison, we detected significant PAX9 protein expression with varying frequency in all the six panels of cancer cell lines that represented SCLC, NSCLC, mesothelioma, head and neck, esophageal and pancreatic cancers. Our studies clearly point out the fact that any given cancer is likely to express one or more PAX genes and PAX5 expression, however, appears to be specific for SCLCs.
In diffuse large B cell lymphomas and lymphoplasmacytoid lymphoma, a subtype of B cell non-Hodgkin's lymphoma, frequent translocation of the PAX5 gene to the IgH chain gene locus [t(9;14) (p13; q32)] was reported, most likely because of aberrant somatic hypermutation events.36, 37 It results in a juxtaposition placement of PAX5 to IgH chain regulatory elements, thereby resulting in a deregulated expression of PAX5. We have used the FISH method that previously identified reproducible PAX15, 16 and non-PAX gene rearrangements38, 39 in other cancers. In this study, based on FISH analysis, we did not find significant PAX5 gene amplification, and there were also no chromosomal translocations in more than 30 tumor tissues examined (Figure 3). In addition, unlike c-Met that acquire gain-of-function mutations in SCLC,12 we failed to detect any significant mutations in PAX5 genomic DNA in SCLC tumor tissues (Figure 4).
RTKs play a very important role in tumor development and metastasis. Based on our previously published data12 and the data shown in Figures 1 and 2, it is clear that the RTK c-Met is highly expressed in SCLC primary tumors and cell lines. Although we failed to detect PAX3 in SCLC, there is still a possibility that PAX5 can activate c-Met transcription because the PAX5 nuclear binding sequence is very similar to that of PAX3 (−476 to −460 upstream of the first non-encoding exon of the human MET gene),11 and c-Met promoter sequence has one putative PAX5 binding site. Using luciferase reporter and ChIP assays, we have shown unequivocally that PAX5, also directly and positively regulates c-Met transcription (Figure 5). The physiological relevance can be appreciated by the fact that knockdown of PAX5 in SCLC cells resulted in a substantial loss in c-Met protein expression, thereby linking directly the expression of c-Met to PAX5. The fact that the PAX5 knockdown cells still express other PAX genes such as PAX2 and 9 shows that PAX5 is the main regulator of c-Met in SCLC cells. We also reported in this as well as in an earlier study that c-Met is also over expressed in NSCLC40, 41 and obviously they lack the detectable PAX5 expression. Therefore, it is likely that the NSCLC tumors regulate their c-Met expression through PAX8 and PAX2, both of which are known paralogs of PAX5; however, this possibility remains to be investigated. Previous work by others identified multiple SP1 binding sites that are likely to aid in the constitutive expression of the c-Met gene.42 PAX5 is also known to repress p53 expression.43 In the untranslated first exon of TP53, there is a PAX5 binding site that is responsible for PAX5-mediated repression of p53 transcription. As most of the SCLC has mutated p53 that makes it non-functional, the above role of PAX5 may not be relevant; however, in those tumors that harbor wt p53, depending on the expression, PAX5 (or PAX2 and PAX8) is very likely to interfere with p53 transcription and, thereby contribute to the tumor development. Daxx, originally identified as Fas cytoplasmic tail binding protein, is known to be localized both in the cytoplasm and the nucleus. In the nucleus Daxx interacts with PAX5 and through HDAC2, appears to have a significant suppressive effect on c-Met transcription.44 The role of Daxx in the context of PAX5 in SCLC cells needs to be investigated.
The PAX transcription factor activities are also regulated by specific protein–protein interactions. For instance, PAX 2, 5 and 8 are known to interact with the tumor suppressor pRB, and in case of PAX8, it has been shown to activate the transcriptional activity.45 Daxx, on the other hand, interacts with PAX5; however, it appears to suppress the activity through its binding to HDAC2.45 The fact that in HGF-treated cells, PAX5 and activated c-Met both co-localize to the same intra-nuclear compartment, adds yet another dimension to the possible PAX5 functions. The role of nuclear phospho-c-Met in these HGF-treated SCLC cells needs to be established. The fact that we were able to successfully co-precipitate c-Met with PAX5 suggests a strong functional link between the two (Figure 6). We have shown previously that c-Met is mutated frequently in SCLC with a gain of function.12 It is possible that in normal cells, nuclear c-Met binds to PAX5 and acts as a feedback inhibitor of PAX5 transcriptional activity; however, in SCLC cells, although the mutated c-Met is translocated into the nucleus in HGF-treated cells, its ability to inhibit PAX5 function may be lost and therefore led to the elevated levels of c-Met in SCLC.
Abrogation of PAX5 is known to decrease viability of cancer cells,6 thereby, suggesting that PAX5 is required for survival and proliferation. It is interesting to note that the PAX5 is expressed in a subset of neuroblastoma cells named N-type but not in the more benign S-type cells.6 However, forced expression of PAX5 in the S-type cells increases their oncogenic potential as evidenced from enhanced cell viability and anchorage independent colony formation. We have also shown in this paper, a significant role for PAX5 in promoting SCLC cell viability. The effect is rather modest, most likely due to the fact that these cells are also expressing other PAX genes such PAX2 and 8 that are known to contribute to the overall survival of the SCLC cells.46 As the prognosis for SCLC is extremely poor, targeted combinatorial therapy is likely to be highly efficacious. Therefore, we tested the effect of PAX5 knockdown in SCLC cells on their viability in combination with c-Met and Topoisomerase 1 inhibitors that are currently used in SCLC therapy.7, 47 The potential adjunct therapeutic role of targeting PAX5 is apparent in the results presented in Figure 7. Loss of PAX5 clearly decreases the viability of SCLC cells; however, the decrease is much greater when combined with c-Met or Topo1 inhibitor; and the maximum effect is seen when all the three conditions are combined. This study revealed one very important aspect in cancers that is the relatively high possibility of multiple PAX genes being expressed simultaneously. Although our study provides the proof of principle with respect to PAX5, in order to get dramatic effects, we need to abrogate the expressions of all the PAX genes and then apply the above combinatorial therapy, an aspect we are investigating currently.
In conclusion, many types of cancers express one or more of PAX proteins. The expression of PAX5 is highly restricted to SCLC and, in comparison, PAX8 is expressed in NSCLC. SCLC has an extremely poor prognosis and as PAX5 transcription factor appears to be expressed preferentially in SCLC, and also because it appears to be directly responsible for the elevated levels of c-Met commonly encountered in SCLC, we propose that PAX5 be targeted for therapeutic purposes.
References
Abidoye O, Ferguson MK, Salgia R . Lung carcinoma in African Americans. Nat Clin Pract Oncol 2007;4:118–129.
Robson EJD, He S-J, Eccles MR . A panorama of PAX genes in cancer and development. Nat Rev Cancer 2006;6:52–62.
Cobaleda C, Schebesta A, Delogu A, et al. Pax5: the guardian of B cell identity and function. Nat Immunol 2007;8:463–470.
Krenacs L, Himmelmann AW, Quintanilla-Martinez L, et al. Transcription factor B-cell-specific activator protein (BSAP) is differentially expressed in B cells and in subsets of B-cell lymphomas. Blood 1998;A92:1308–1316.
Kozmik Z, Sure U, Rüedi D, et al. Deregulated expression of PAX5 in medulloblastoma. Proc Natl Acad Sci USA 1995;92:5709–5713.
Baumann Kubetzko FB, Di Paolo C, Maag C, et al. The PAX5 oncogene is expressed in N-type neuroblastoma cells and increases tumorigenicity of a S-type cell line. Carcinogenesis 2004;25:1839–1846.
Sattler M, Salgia R . c-Met and hepatocyte growth factor: potential as novel targets in cancer therapy. Curr Oncol Rep 2007;9:102–108.
Rygaard K, Nakamura T, Spang-Thomsen M . Expression of the proto-oncogenes c-met and c-kit and their ligands, hepatocyte growth factor/scatter factor and stem cell factor, in SCLC cell lines and xenografts. Br J Cancer 1993;67:37–46.
Maulik G, Kijima T, Ma PC, et al. Modulation of the c-Met/hepatocyte growth factor pathway in small cell lung cancer. Clin Cancer Res 2002;8:620–627.
Olivero M, Rizzo M, Madeddu R, et al. Overexpression and activation of hepatocyte growth factor/scatter factor in human non-small-cell lung carcinomas. Br J Cancer 1996;74:1862–1868.
Epstein JA, Shapiro DN, Cheng J, et al. Pax3 modulates expression of the c-Met receptor during limb muscle development. Proc Natl Acad Sci USA 1996;93:4213–4218.
Ma PC, Jagadeeswaran R, Jagadeesh S, et al. Functional expression and mutations of c-Met and its therapeutic inhibition with SU11274 and small interfering RNA in non-small cell lung cancer. Cancer Res 2005;65:1479–1488.
Fujita N, Wade PA . Use of bifunctional cross-linking reagents in mapping genomic distribution of chromatin remodeling complexes. Methods 2004;33:81–85.
Sato Y, Sugie R, Tsuchiya B, et al. Comparison of the DNA extraction methods for polymerase chain reaction amplification from formalin-fixed and paraffin-embedded tissues. Diagn Mol Pathol 2001;10: 265–271.
French CA, Alexander EK, Cibas ES, et al. Genetic and biological subgroups of low-stage follicular thyroid cancer. Am J Pathol 2003;162:1053–1060.
Kroll TG, Sarraf P, Pecciarini L, et al. PAX8-PPARg1 fusion oncogene in human thyroid carcinoma [corrected]. Science 2000;289:1357–1360.
Relaix F, Polimeni M, Rocancourt D, et al. The transcriptional activator PAX3-FKHR rescues the defects of Pax3 mutant mice but induces a myogenic gain-of-function phenotype with ligand-independent activation of Met signaling in vivo. Genes Dev 2003;17:2950–2965.
Gomes DA, Rodrigues MA, Leite MF, et al. c-Met must translocate to the nucleus to initiate calcium signals. J Biol Chem 2008;283:4344–4351.
Pozner-Moulis S, Pappas DJ, Rimm DL . Met, the hepatocyte growth factor receptor, localizes to the nucleus in cells at low density. Cancer Res 2006;66:7976–7982.
Salgia R . c-Met inhibition. Clin Adv Hematol Oncol 2006;4:823–824.
Krenacs L, Himmelmann AW, Quintanilla-Martinez L, et al. Transcription factor B-cell-specific activator protein (BSAP) is differentially expressed in B cells and in subsets of B-cell lymphomas. Blood 1998;92:1308–1316.
Foss HD, Reusch R, Demel G, et al. Frequent expression of the B-cell-specific activator protein in Reed-Sternberg cells of classical Hodgkin's disease provides further evidence for its B-cell origin. Blood 1999;94:3108–3113.
Sica G, Vazquez MF, Altorki N, et al. PAX5 expression in pulmonary neuroendocrine neoplasms: its usefulness in surgical and fine-needle aspiration biopsy specimens. Am J Clin Pathol 2008;129: 556–562.
Mhawech-Fauceglia P, Saxena R, Zhang S, et al. PAX5 immunoexpression in various types of benign and malignant tumours: a high-throughput tissue microarray analysis. J Clin Pathol 2007;60:709–714.
Poleev A, Fickenscher H, Mundlos S, et al. AX8, a human paired box gene: isolation and expression in developing thyroid, kidney and Wilms’ tumors. Development 1992;116:611–623.
Tagge EP, Hanson P, Re GG, et al. Paired box gene expression in Wilms’ tumor. J Pediatr Surg 1994;29:134–141.
Eccles MR, Yun K, Reeve AE, et al. Comparative in situ hybridization analysis of PAX2, PAX8, and WT1 gene transcription in human fetal kidney and Wilms’ tumors. Am J Pathol 1995;146:40–45.
Bouchard M, Souabni A, Mandler M, et al. Nephric lineage specification by Pax2 and Pax8. Genes Dev 2002;16:2958–2970.
Lui WO, Foukakis T, Lidén J, et al. Expression profiling reveals a distinct transcription signature in follicular thyroid carcinomas with a PAX8-PPAR (gamma) fusion oncogene. Oncogene 2005;24:1467–1476.
Scouten WT, Patel A, Terrell R, et al. Cytoplasmic localization of the paired box gene, Pax-8, is found in pediatric thyroid cancer and may be associated with a greater risk of recurrence. Thyroid 2004;14:1037–1046.
Shapiro DN, Sublett JE, Li B, et al. Fusion of PAX3 to a member of the forkhead family of transcription factors in human alveolar rhabdomyosarcoma. Cancer Res 1993;53:5108–5112.
Galili N, Davis RJ, Fredericks WJ, et al. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat Genet 1993;5:230–235.
Barr FG, Galili N, Holick J, et al. Rearrangement of the PAX3 paired box gene in the paediatric solid tumour alveolar rhabdomyosarcoma. Nat Genet 1993;3:113–117.
Anderson J, Ramsay A, Gould S, et al. PAX3-FKHR induces morphological change and enhances cellular proliferation and invasion in rhabdomyosarcoma. Am J Pathol 2001;159:1089–1096.
Gerber JK, Richter T, Kremmer E, et al. Progressive loss of PAX9 expression correlates with increasing malignancy of dysplastic and cancerous epithelium of the human oesophagus. J Pathol 2002;197:293–297.
Poppe B, De Paepe P, Michaux L, et al. PAX5/IGH rearrangement is a recurrent finding in a subset of aggressive B-NHL with complex chromosomal rearrangements. Genes Chromosomes Cancer 2005;44:218–223.
Ohno H, Ueda C, Akasaka T . The t (9;14)(p13;q32) translocation in B-cell non-Hodgkin's lymphoma. Leuk Lymphoma 2000;36:435–4345.
Perner S, Demichelis F, Beroukhim R, et al. TMPRSS2:ERG fusion-associated deletions provide insight into the heterogeneity of prostate cancer. Cancer Res 2006;66:8337–8341.
Tomlins SA, Rhodes DR, Perner S, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science 2005;310:644–648.
Ma PC, Schaefer E, Christensen JG, et al. A selective small molecule c-MET Inhibitor, PHA665752, cooperates with rapamycin. Clin Cancer Res 2005;11:2312–2319.
Jagadeeswaran R, Jagadeeswaran S, Bindokas VP, et al. Activation of HGF/c-Met pathway contributes to the reactive oxygen species generation and motility of small cell lung cancer cells. Am J Physiol Lung Cell Mol Physiol 2007;292:L1488–L1494.
Zhang X, Li Y, Dai C, et al. Sp1 and Sp3 transcription factors synergistically regulate HGF receptor gene expression in kidney. Am J Physiol Renal Physiol 2003;284:F82–F94.
Stuart ET, Haffner R, Oren M, et al. Loss of p53 function through PAX-mediated transcriptional repression. EMBO J 1995;14:5638–5645.
Emelyanov AV, Kovac CR, et al. The interaction of Pax5 (BSAP) with Daxx can result in transcriptional activation in B cells. J Biol Chem 2002;277:11156–11164.
Miccadei S, Provenzano C, Mojzisek M, et al. Retinoblastoma protein acts as Pax 8 transcriptional coactivator. Oncogene 2005;24: 6993–7001.
Park D, Jia H, Rajakumar V, et al. Pax2/5/8 proteins promote cell survival in C. elegans. Development 2006;133:4193–4202.
Lally BE, Urbanic JJ, Blackstock AW, et al. Small cell lung cancer: have we made any progress over the last 25 years? Oncologist 2007;12:1096–1104.
Acknowledgements
We would like to cordially thank Dr Elizabeth Hyjek for her assistance with PAX5 immunohistochemical verification on diagnostic control cases. Grants from NIH/NCI, American Lung Association, Mesothelioma Applied Research Foundation (Jeffrey P Hayes Award), and V-Foundation (Guy Geleerd Memorial Award) awarded to Ravi Salgia supported the work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Kanteti, R., Nallasura, V., Loganathan, S. et al. PAX5 is expressed in small-cell lung cancer and positively regulates c-Met transcription. Lab Invest 89, 301–314 (2009). https://doi.org/10.1038/labinvest.2008.168
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/labinvest.2008.168
Keywords
This article is cited by
-
Polymorphism in lncRNA AC016683.6 and its interaction with smoking exposure on the susceptibility of lung cancer
Cancer Cell International (2018)
-
PI3 Kinase Pathway and MET Inhibition is Efficacious in Malignant Pleural Mesothelioma
Scientific Reports (2016)
-
Pulmonale neuroendokrine Tumoren in der neuen WHO-Klassifikation 2015
Der Pathologe (2015)
-
Role of PAX8 in the regulation of MET and RON receptor tyrosine kinases in non-small cell lung cancer
BMC Cancer (2014)
-
Therapeutic implications of activation of the host gene (Dleu2) promoter for miR-15a/16-1 in chronic lymphocytic leukemia
Oncogene (2014)