Abstract
EGFR mutation-induced drug resistance has significantly impaired the potency of small molecule tyrosine kinase inhibitors in lung cancer treatment. Computational approaches can provide powerful and efficient techniques in the investigation of drug resistance. In our work, the EGFR mutation feature is characterized by the energy components of binding free energy (concerning the mutant-inhibitor complex) and we combine it with specific personal features for 168 clinical subjects to construct a personalized drug resistance prediction model. The 3D structure of an EGFR mutant is computationally predicted from its protein sequence, after which the dynamics of the bound mutant-inhibitor complex is simulated via AMBER and the binding free energy of the complex is calculated based on the dynamics. The utilization of extreme learning machines and leave-one-out cross-validation promises a successful identification of resistant subjects with high accuracy. Overall, our study demonstrates advantages in the development of personalized medicine/therapy design and innovative drug discovery.
Similar content being viewed by others
Introduction
Non-small-cell lung cancer (NSCLC) has become a major threat to human health1. Mutations, such as in-frame deletions or amino acid substitutions, clustered around the ATP-binding pockets of the tyrosine kinase domain of the epidermal growth factor receptor (EGFR) are the primary cause of NSCLC1,2,3. In clinical treatment of NSCLC, tyrosine kinase inhibitors (TKIs) such as gefitinib and erlotinib are widely used3,4. These two reversible inhibitors show stronger binding affinity with mutant kinases than the wild-type (WT) EGFR and they indeed produce good results for many patients for a period of time2.
However, the effectiveness of these inhibitors is limited by the emergence of drug resistance, sometimes due to a second mutation, such as the substitution of threonine with methionine at residue site 7902,3. The cause of drug resistance is thought to be steric interference with the binding of inhibitors caused by the mutations5,6,7. Irreversible inhibitors including CL387/785, EKB-569 and HKI-272 are proposed to tackle the problem5,6,8,9,10. However, the EGFR structure will be chemically modified via a covalent bond2, which is not encouraged in practical therapy. Therefore, the EGFR mutation-induced drug resistance leads to an urgent demand to develop new treatment strategies11,12.
With the rapid development of bioinformatics, computational methods13,14 have become more efficient and popular for studying the molecular mechanism of mutation-induced drug resistance, developing predictive tools and designing resistance-evading drugs4,11,12,15. These computational approaches are investigated based on the genotypic data, which fall into two categories: sequence-based and structure-based approaches. With the utilization of three-dimensional (3D) structural information16, machine learning and pattern classification methods such as neural networks17,18,19, support vector machines (SVM)20 and decision trees21 have shown high potential in the prediction of drug resistance and innovative drug design11.
In this paper, we present a method that combines the EGFR-inhibitor interaction pattern and the specific personal features for each of our 168 clinical subjects to construct a personalized drug resistance prediction model. Our method can have useful applications to the development of personalized medicine/therapy. In this method, mutations in protein sequences of the EGFR kinase domain are initially translated into the 3D structures based on a template structure, using protein structure prediction tools scap22 and loopy23. AMBER24 is employed to simulate the dynamics of the kinase mutant-inhibitor systems and evaluate the binding free energies of the mutants and inhibitors. We then characterize the EGFR-inhibitor interaction by the energy components of the binding free energy extracted via MM/PBSA in AMBER24. These interaction patterns coupled with specific personal features of our subjects are regarded as main characteristics for further classification. Extreme learning machines (ELMs)25,26 are adopted here together with leave-one-out cross-validation. These structural analyses provide us with insights into the mechanism of mutation-induced drug resistance at the molecular level, which play an important role in personalized therapy design and innovative drug discovery.
Results
Inhibitors
Gefitinib (IRESSA™) and erlotinib (TARCEVA®) are the main inhibitors used in EGFR-targeted therapy. We isolate them from their bound complexes 2ITY and 1M17 downloaded from the Protein Data Bank (PDB)16. Their 3D structures can be viewed in Figure 1 (parts a and b). The General AMBER Force Field (GAFF), which covers most of the organic chemical space, is implemented to generate the topology and coordinate files of the inhibitors. Based on GAFF, the antechamber program in AMBER24 assigns atomic charges and atom/bond types for the inhibitors and further constructs their topology files. The AM1-BCC charge method27, which efficiently reproduces the HF/6-31G* RESP charge, is employed when adding atomic charges.
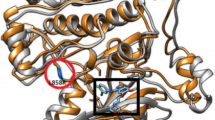
3D structures of inhibitors, computationally predicted mutants and complexes.
Parts (a) and (b) show the 3D structures of inhibitors gefitinib (IRESSA™) and erlotinib (TARCEVA®) respectively. In parts (c) to (g), we present a comparison between the mutation neighborhood of our computationally predicted mutant and the corresponding site of the WT EGFR kinase protein, for a specific mutation type. Each white chain corresponds to the WT structure and each blue one is our modeling result. Accordingly, parts (c) to (g) show the mutation types L858R, delL747_P753insS, dulH773, delE746_A750 and T854A_L858R respectively. Parts (h) and (i) display the inhibitor-binding pocket of mutant delE746_A750 with inhibitors gefitinib and erlotinib respectively.
Results for the modeling of mutant-inhibitor complexes
In our study, we focus on the mutations on exons 18 ~ 21 of the EGFR tyrosine kinase domain. Specifically, we carried out clinical observations on 168 lung-cancer patients from the Queen Mary Hospital in Hong Kong. These patients are then mapped from their genotypes into a total of 37 mutation types of the WT EGFR kinase protein. We notate these mutation types by their corresponding changes in protein sequences relative to the WT sequence, as the following principles (refer to Supplementary Table 1 for an overall list).
-
Residue substitution of X with Y at residue site I is denoted by XIY, such as L858R.
-
Deletion of residues at sites I (residue X) to II (residue Y) is denoted by delXI_YII, such as delE746_A750.
-
Duplication of residues at sites I (residue X) to II (residue Y) is represented as dulXI_YII, such as dulS768_D770.
-
Modification of residues at sites I (residue X) to II (residue Y) is denoted by a combination of deletion and insertion (delXI_YIIinsk, k is a residue list), such as delL747_A755insSKG.
-
A double-point mutation of X with Y at residue site I and A with B at residue site II is named by two single-point mutations connected by an underscore, such as T854A_L858R.
Further, we carry out statistics for these mutation types on our patients and derive that mutation types L858R (80 cases), delE746_A750 (38 cases) and delL747_P753insS (10 case) occupy the majority of the patients, while the others are considered as rare mutations. For simplicity in our later interpretation, we name the mutants the same as their corresponding mutation types, such as mutant L858R and mutation type L858R.
Subsequently, we translate these mutations from protein sequences into their 3D structures. A mutated protein structure is determined based on homology modeling. Different types of mutations are then obtained using two programs, scap22 and loopy23. First, the template protein structures extracted from complex 2ITZ and 2ITY are prepared. Scap deals with side chain substitutions. It packs side chains that are selected from a previously constructed rotamer library28, according to the energy preferences coupled with steric feasibility29. Meanwhile, loopy handles both residue deletion and insertion. The core of loopy is the solution of a mini protein folding problem. Accordingly, it samples the conformation space with constraints of closure30 and steric feasibility29 and scores the candidates based on the colony energy23. Some examples of the modeling results are displayed in Figure 1 (parts c to g). The 3D structures are displayed using UCSF Chimera31. For each sampled structure we carry out a rough minimization32, where the maximum number of minimization steps is set as 5000 with the first 2500 steps performed using the steepest descent algorithm. Inhibitors (gefitinib and erlotinib) are separately aligned to the binding pocket of each mutant structure, to construct their bound complexes. As an example, the binding pocket of mutant delE746_A750 for gefitinib and erlotinib is exhibited in Figure 1 (parts h and i).
Furthermore, for the three dominant mutation types from our observed patients, namely L858R, delE746_A750 and delL747_P753insS, we carry out a brief exploration in Figure 2 on the modeled mutant-inhibitor complex structures, with the WT-inhibitor system used for a comparison. In this figure, we comparably display the inhibitor-binding pocket and mutation site of each mutant and those sites of the WT protein. We can see that, the frequently mutated sites are located in the loops at the margin or neighborhood of the inhibitor-binding pocket. It is well acknowledged that, loops23,29 are more flexible than other protein secondary structures, such as α–helixes and β-sheets33, which to some extent explains why these mutations occur easily and frequently in the WT structure. A comprehensive survey in the future will provide deeper insights into these structures.
A comparison between the mutant-inhibitor complex and the WT-inhibitor complex structures for several major mutation types.
In each diagram, a portion of a WT/mutant-inhibitor complex is presented, with the inhibitor (gefitinib) colored pink and the original/mutation site colored blue. Diagrams (a) and (b) show a comparison between the WT-gefitinib system and the L858R-gefitinib system. Similarly, diagrams (c) ~ (d) and (e) ~ (f) show mutations delL747_P753insS and delE746_A750 respectively.
Molecular dynamics (MD) simulations
Each acquired mutant-inhibitor complex is then computationally solvated into a water box. The dynamics of the complex is simulated in this solvent environment. Prior to the crucial MD simulation, the entire system should be equilibrated to a stable state. We employ sander in AMBER for a series of equilibrating operations, which incorporates a short 1000-step minimization (the first half with the steepest descent steps) to remove bad contacts, a 50-picosecond (ps) heating (0 ~ 300 K) and a 50 ps density equilibration with weak restraints (weight of 2.0) from a harmonic potential on the mutant-inhibitor complex and a 500 ps constant pressure equilibration at 300 K. All simulations are performed with SHAKE constraints on hydrogen atoms to remove their bond stretching freedom and the Langevin dynamics is adopted for an efficient temperature control. The equilibration of each system is verified through observing the temperature, density, energy and backbone root-mean-square deviation (RMSD) of each system.
Once each system equilibration is achieved, we generate the production MD simulation for 2 nanoseconds (ns), where we collect trajectory frames at a step of 10 ps and 200 frames in each trajectory. A stable backbone RMSD in each system is an apparent indicator of the stabilization of the production MD simulation, which guarantees a posterior reliable calculation of the binding free energy. For each system, the backbone RMSD distribution over the simulation period (2 ns) is investigated. As an example, the plots for trajectory vs. backbone RMSD in this period, with regard to several major systems, are shown in Figure 3. These systems each incorporate an EGFR kinase protein (WT, L858R, delE746_A750 or delL747_P753insS) and an inhibitor (gefitinib or erlotinib). In this figure, the backbone RMSD values show an acceptable level of stabilization for each system.
An investigation of the stabilization of several solvated mutant/WT-inhibitor systems.
Diagrams (a) and (b) show the plots for trajectory (frames) vs. backbone RMSD (Å) in the MD simulation period (2 ns), with regard to the solvated WT-gefitinib and WT-erlotinib systems respectively. Similarly, diagrams (c) ~ (d), (e) ~ (f) and (g) ~ (h) present the plots for the systems involving L858R, delE746_A750 and delL747_P753insS respectively.
Binding free energy
The production MD simulations produce the motion trajectories of the solvated mutant-inhibitor systems and the binding free energies are calculated based on these trajectories. Binding free energy is a quantitative estimate of the binding affinity of a solvated receptor-ligand system. Based on the computations of different types of free energy differences, MMPBSA in AMBER derives the binding free energies, which encompass energy components of Van der Waals forces (VDW), electrostatic interactions (EEL) and the polar (EPB) and non-polar (ENPOLAR) terms of the solvation free energies. For the WT protein and observed mutants, we calculate their binding free energies with the two inhibitors gefitinib and erlotinib respectively. The detailed information of these energies and their components can be referred to in Supplementary Table 1.
We further examine the distributions of these obtained binding free energies and their components (VDW, EEL, EPB and ENPOLAR) in Figure 4 (parts b to e). The distribution of the binding free energy of these mutants (with WT protein included) with gefinitinib is displayed in part b of Figure 4 and that with erlotinib involved is shown in part d. For both inhibitors gefitinib and erlotinib, the binding free energies with these mutants scatter around that with the WT protein (denoted by red lines). Especially, for mutation L858R that is a common cause of lung cancer, the binding free energy of the mutant with an inhibitor (marked with solid blue circles) is lower than that of the WT protein. In parts c and e of Figure 4, we give the distributions of the binding free energy components coupled with that of the total energy, separately concerning the two inhibitors. The extracted energy components VDW, EEL, EPB and ENPOLAR possess different distributions to the total energy, which may reveal potential significant features for these mutation types. On the other hand, we display the statistics for the mutation types on our 168 patients in part a of Figure 4, where the three peaks are L858R, delE746_A750 and delL747_P753insS respectively, as aforementioned.
Statistics on mutation types and their binding free energies with the two inhibitors.
Part (a) shows the statistics of the 37 mutation types of our observed 168 patients. Parts (b) and (d) present the distributions of total binding free energies of the mutants (with WT protein included) with two inhibitors gefitinib and erlotinib. The red lines and solid blue circles show the binding free energy for the WT EGFR and the L858R mutant respectively. Parts (c) and (e) display the distributions of the binding free energy components, which encompass VDW, EEL, EPB and ENPOLAR, for the two inhibitors.
Computational prediction of drug resistance
The potency of an inhibitor in the treatment of a specific patient can be measured by its survival time or response level. In clinical observations, survival time is generally recorded in unit of months or days, corresponding to a continuous variable in computation. Response level can be divided into four categories and thus mapped into a discrete variable ranging in [1, 4]. Each of our 168 patients has been clinically observed and recorded by RECIST in his treatment involving a specific inhibitor (gefinitib or erlotinib).
Firstly, we simply examine how the computed binding free energy (total energy) relates to the survival time or response level. For a specific patient, the feature of binding free energy (and energy components) can be derived from his/her EGFR mutation type combined with the inhibitor used in his/her treatment (checked in Supplementary Table 1). Gefitinib is applied in majority of the treatments for the 168 patients (137 cases of gefitinib, 31 cases of erlotinib). To normalize the potency of the two inhibitors (gefinitib and erlotinib), we set the binding free energies of the WT protein with the two inhibitors as baselines, which implies that for each case concerning a mutation type and an inhibitor we subtract the baseline value from its binding free energy (same for the components) to obtain the final energy-related feature. We call the energy-related feature “mutation feature” for short in the following interpretation. In part a of Figure 5, we plot the distribution of mutation feature (which represents the total binding free energy) vs. survival time and that of mutation feature vs. response level is displayed in part b. From parts a and b of Figure 5, we find that the mutation feature is not one-to-one related or linearly related to the survival time or response level, which demonstrates the influence of individual difference on the potency of an inhibitor.
Statistics on features and classification results of the clinical subjects.
Part (a) shows the distribution of mutation feature (total binding free energy) vs. survival time for the 168 clinical subjects, with each point representing one subject. Similarly, the plot of mutation feature vs. response level is displayed in part (b). The distributions of the adopted features (personal + mutation) for the 168 subjects are shown in parts (c) and (d), with part (c) showing the original distribution while part (d) the normalized values. Part (e) provides a comparison between the training accuracies reported in the case involving the mutation feature only (blue, denoted as ‘M’) and the case involving both mutation feature and personal features (brown, denoted as ‘M + P’). Part (f) shows a comparison between the testing accuracies (blue for the first case and brown for the second).
Personal information of each patient is recorded as well. The personal information, which is referred to as “personal features” in later interpretation, incorporates basic descriptive features and symptoms. Detailed items include age, gender, smoking history, performance status, subtypes of the NSCLC, stages describing the development of the NSCLC, brain metastasis and suspension of TKIs. For simplicity, we further discretize the original age values into ranges (0, 50), [50, 60), [60, 70), [70, 80), [80, 200), which is finally mapped into a discrete range of [0, 4]. Detailed information for all these personal features is provided in Table 1.
We next combine the personal features with the mutation feature for each patient, leading to a total of 168 subjects, to develop a drug resistance prediction model (to predict response level). Before building the model, we normalize each feature of the whole feature set (15 features) into a range of [−1, 1] to compensate the differences. The distributions of values of these features (personal + mutation) for the 168 subjects are shown in parts c and d of Figure 5. Part c gives the original distribution while part d exhibits the normalized values.
Extreme learning machines (ELMs) are used for building a classification model, in which an optimal set of weights and biases are determined by finding a least-square solution with a previously calculated hidden layer output matrix. With the generally used sigmoidal function applied as the activation function g(x), the required number of hidden nodes  is regarded as the single controllable parameter. Our validation system is the leave-one-out cross-validation, in which we build the classifier 168 times and a different sample is used for testing in turn each time with the remaining 167 samples used for training. The required number of hidden nodes
is regarded as the single controllable parameter. Our validation system is the leave-one-out cross-validation, in which we build the classifier 168 times and a different sample is used for testing in turn each time with the remaining 167 samples used for training. The required number of hidden nodes  in ELMs is adapted from 50 to 500 at a step of 50. For each specific
in ELMs is adapted from 50 to 500 at a step of 50. For each specific  , the ELM will be repeated 20 times and the best performance is retained. The final classification accuracy is calculated by averaging the classification rates of all 168 classifiers. In order to conduct a comparison between the case where only the mutation feature is used and the case involving both the mutation feature and personal features, we apply the classification model on both these two cases and the results are presented in Table 2. As shown in this table, with
, the ELM will be repeated 20 times and the best performance is retained. The final classification accuracy is calculated by averaging the classification rates of all 168 classifiers. In order to conduct a comparison between the case where only the mutation feature is used and the case involving both the mutation feature and personal features, we apply the classification model on both these two cases and the results are presented in Table 2. As shown in this table, with  ranging from 50 to 500, we obtain average classification accuracies of 81.55% and 68.33% for training and testing respectively in the first case (using the mutation feature alone); while 95.41% and 89.94% are achieved in the second case (using both mutation and personal features). This implies the necessity of incorporating personal features into the model and a detailed comparison between the training/testing accuracies reported in the two cases is shown in Figure 5 (parts e and f). Furthermore, the best testing accuracy 95.83% (for the second case) is achieved with
ranging from 50 to 500, we obtain average classification accuracies of 81.55% and 68.33% for training and testing respectively in the first case (using the mutation feature alone); while 95.41% and 89.94% are achieved in the second case (using both mutation and personal features). This implies the necessity of incorporating personal features into the model and a detailed comparison between the training/testing accuracies reported in the two cases is shown in Figure 5 (parts e and f). Furthermore, the best testing accuracy 95.83% (for the second case) is achieved with  = 150, which reflects a very good prediction result.
= 150, which reflects a very good prediction result.
Discussion
The field of bioinformatics is developing very rapidly and it makes the prediction of molecular structure, studies of mutation-induced drug resistance and innovative drug discovery more feasible4,11,12. In this work, we develop a computational model to transfer the genotypic data to phenotypic data for specialized subjects, by characterizing the EGFR-inhibitor interaction patterns and taking personal features into consideration. The constructed mutant-inhibitor complexes are each solvated into a solvent environment and a successive systematic equilibrium is achieved via simulations. We subsequently characterize the features of a subject using the energy components of binding free energy (mutation feature) and the specialized personal information (personal features). The combination of ELMs and leave-one-out cross-validation produces a successful identification of resistant subjects with high accuracy.
Personalized medicine/therapy proposes customization of healthcare to individual patients and the use of genotypic information plays an important role. Our method can be regarded as a personalized prediction model for drug resistance, based on both the mutation feature and the personal features of a patient. With a high prediction rate for drug resistance, our model encourages the development of personalized medicine/therapy design.
As one of our future works, more accurate and powerful approaches will be explored to predict the 3D structure of a specific mutation, based on its sequential information. Homology modeling will serve as a guiding role34,35. On the other hand, more efficient strategies will be discovered to reduce the high computational complexity of calculating the binding free energy for a mutant-inhibitor system. Modern graphics processing units (GPUs) and field programmable gate arrays (FPGAs) have evolved into high performance accelerators for parallel computing36,37,38,39. With these devices, the computational power can be improved tens or even hundreds of times. In our future computations, GPUs and FPGAs will be adopted to accelerate the computation of binding free energies. Moreover, the binding free energy library (Supplementary Table 1) can be periodically updated so that only newly identified mutation types need to be added to the library. Since the mutation types in a dataset are highly redundant, the utilization of this library will significantly reduce the computational load. Thus, more clinical data can be collected and analyzed in our following studies, which will improve the aforementioned prediction model and help us update the library based on the new data. Future studies will bring more benefits to the investigation of mutation-induced drug resistance and innovative drug design.
Methods
EGFR kinase mutant-inhibitor complex modeling and molecular dynamics (MD) simulations
First, we predict the structures of EGFR kinase domain mutations computationally. The scap program handles side chain substitutions and the loopy program handles residue deletions and insertions. After obtaining the predicted structures, we optimize them through the QM/MM mechanism in AMBER. Missing atoms and an octahedron water box (10 angstrom) are added using the tleap program before we carry out minimization with the sander program. Each structure is initially partitioned into a QM region for the mutated residues and an MM region for the other residues and the system is characterized by an effective Hamiltonian as described in Equation (1).

Here the MM region is handled classically using the AMBER additive force field (Equation (2)); the QM region and QM/MM interface are formulated with Hamiltonians.

Once a refined mutant structure is obtained, we align it to the template complex 2ITY or 1M17 containing the WT kinase protein and the drug molecule, to acquire an original coarse mutant-inhibitor complex. Likewise, we use AMBER to minimize these complexes and simulate their dynamics in a solvent environment. AMBER adopts Equation (2) as the basic force field form during molecular dynamics (MD) simulations and the ff99SB force field is selected in our work owing to its broad applications. A simple water box with a 10.0 angstrom buffer around the complex in each direction is generated, based on the common TIP3P water model. The tleap program creates the topology and coordinates files of the solvated complex and passes them to sander for the later MD run.
An important factor for achieving a stable MD simulation is the equilibrium of a system. Using AMBER and AMBER TOOLS, we build up a moderate setting for the equilibration, which encompass minimization, heating, density equilibration and constant pressure equilibration, leading to an approximately 4-hour run on 12 3.47 GHz processors of our computer. The subsequent MD simulation is performed on each equilibrated system with a relatively short time of 2 ns, which aims to compensate for the large computational costs and ultimately leads to a 14-hour run. In the MD run for a mutant-inhibitor complex, the motion trajectory is collected every 10 ps to reach a total of 200 frames, which will be used in the following calculation of the binding free energy.
Molecular binding affinities calculated using MM/PBSA model
The binding free energy of a receptor-ligand complex in a solvent environment is an important standard for measuring the binding affinity. Based on the theory of thermodynamic cycle, the original calculation can be constructed as follows,

Here each ΔG stands for the free energy difference between two distinct states. ΔGBind,Solv and ΔGBind,Vacuum correspond to the free energy difference between the bound and unbound states of a complex in solvent and vacuum respectively. ΔGSolv (ΔGSolv,Ligand, ΔGSolv,Receptor and ΔGSolv,Complex) represents the change of free energy between the solvated and vacuum states of a ligand, receptor or complex.
Both the free energy difference in vacuum ΔGBind,Vacuum and the solvation free energies ΔGSolv contribute to the calculation of binding free energy ΔGBind,Solv. MMPBSA.py in AMBER24 performs Molecular Mechanics/Poisson Boltzmann Surface Area (MM/PBSA) to derive these free energy differences. ΔGBind,Vacuum (Equation (4)) can be captured by averaging the interaction energies ΔE between the receptor and ligand. However, the entropy contribution ΔSNMA is generally neglected for states with similar entropies, due to high computational expense. Practically, energy components for this portion incorporate the Van der Waals forces and the electrostatic interactions between atoms in the MM region. On the other hand, the solvent free energy ΔGSolv typically encompasses the polar contribution and the nonpolar contribution (Equation (5)).


Here, the nonpolar contribution ΔGnonpolar is simply computed by a linear model and the polar portion ΔGpolar is approximated by solving the PB equation.
In this work, a parallel version of MMPBSA.py.MPI is implemented on 12 3.47 GHz processors to accelerate the computations. Each previously obtained MD trajectory, representing a number of conformations, is a major input to MMPBSA. A single MMPBSA run requires 0.5 hours approximately.
Classification models using extreme learning machines
With the personal information (age, gender, smoking history etc.) taken into account, we combine them with the previously acquired energy components of the binding free energy (mutation feature) as principal features and predict the response levels of our observed patients using machine learning techniques.
The fundamental prediction method we adopt is the ELMs. They play an important role in training single-hidden layer feed-forward neural networks (SLFNs) in Equation (6) below and provide a good balance between the computational speed and generalization performance25,26.

Here  is the training set,
is the training set,  and g(x) are the number of hidden nodes and activation function respectively of the SLFN and wj and bj represent the input weights and input biases. The goal is to approximate the training examples with minimum error between oi (Equation (6)) and yi, which could be summarized in a matrix form as Equation (7).
and g(x) are the number of hidden nodes and activation function respectively of the SLFN and wj and bj represent the input weights and input biases. The goal is to approximate the training examples with minimum error between oi (Equation (6)) and yi, which could be summarized in a matrix form as Equation (7).

where H is the hidden layer output matrix. The essential idea of ELMs is to randomly assign the input weights wj and biases bj, which leads the training of the above SLFN to finding a least-square solution  of the linear system denoted by Equation (8).
of the linear system denoted by Equation (8).

The algorithm can be summarized as follows:
Algorithm 1: Extreme Learning Machine
Input:
Training set  which contains N training examples;
which contains N training examples;
Activation function g(x);
Number of hidden node  ;
;
Output:
Input weight wj, input bias bj and output weight β;
-
Randomly assign input weight wj and bias bj where j = 1,…,
 ;
; -
Calculate the hidden layer output matrix H;
-
Calculate the output weight β by β = H†Y, where H† is the Moore-Penrose generalized inverse of matrix H and
 .
.
The generally used sigmoidal function is applied as the activation function g(x) and the required number of hidden nodes  is regarded as a controllable parameter. In addition, when training the model, we employ the leave-one-out cross-validation mechanism, which guarantees each sample is used once as the validation data. The process involves a total number of 168 combinations of partition, training and validation and each combination is referred to as a fold. With a specific parameter setting in each fold, the experiments are repeated 20 times owing to the randomness of ELMs and the best performance will be retained. For the overall cross-validation scheme encompassing 168 folds, we average their results to produce the final one.
is regarded as a controllable parameter. In addition, when training the model, we employ the leave-one-out cross-validation mechanism, which guarantees each sample is used once as the validation data. The process involves a total number of 168 combinations of partition, training and validation and each combination is referred to as a fold. With a specific parameter setting in each fold, the experiments are repeated 20 times owing to the randomness of ELMs and the best performance will be retained. For the overall cross-validation scheme encompassing 168 folds, we average their results to produce the final one.
References
Lynch, T. J. et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med 350, 2129–2139 (2004).
Yun, C. H. et al. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proc Natl Acad Sci U S A 105, 2070–2075 (2008).
Zhang, Z. F. et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet 44, 852–60 (2012).
Hou, T. J., Zhang, W., Wang, J. & Wang, W. Predicting drug resistance of the HIV-1 protease using molecular interaction energy components. Proteins 74, 837–846 (2009).
Kobayashi, S. et al. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. New Engl J Med 352, 786–792 (2005).
Kwak, E. L. et al. Irreversible inhibitors of the EGF receptor may circumvent acquired resistance to gefitinib. P Natl Acad Sci USA 102, 7665–7670 (2005).
Pao, W. et al. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. Plos Med 2, 225–235 (2005).
Carter, T. A. et al. Inhibition of drug-resistant mutants of ABL, KIT and EGF receptor kinases. P Natl Acad Sci USA 102, 11011–11016 (2005).
Greulich, H. et al. Oncogenic transformation by inhibitor-sensitive and -resistant EGFR mutants. Plos Med 2, 1167–1176 (2005).
Sequist, L. V. Second-generation epidermal growth factor receptor tyrosine kinase inhibitors in non-small cell lung cancer. Oncologist 12, 325–330 (2007).
Cao, Z. W. et al. Computer prediction of drug resistance mutations in proteins. Drug Discov Today 10, 521–529 (2005).
Hao, G. F., Yang, G. F. & Zhan, C. G. Structure-based methods for predicting target mutation-induced drug resistance and rational drug design to overcome the problem. Drug Discov Today 17, 1121–1126 (2012).
Sneddon, M. W. & Emonet, T. Modeling cellular signaling: taking space into the computation. Nat Methods 9, 239–242 (2012).
Cohen, A. R., Gomes, F. L., Roysam, B. & Cayouette, M. Computational prediction of neural progenitor cell fates. Nat Methods 7, 213–218 (2010).
Loo, L. H., Wu, L. F. & Altschuler, S. J. Image-based multivariate profiling of drug responses from single cells. Nat Methods 4, 445–453 (2007).
Berman, H. M. et al. The Protein Data Bank. Nucleic Acids Res 28, 235–242 (2000).
Draghici, S. & Potter, R. B. Predicting HIV drug resistance with neural networks. Bioinformatics 19, 98–107 (2003).
Wang, D. C. & Larder, B. Enhanced prediction of lopinavir resistance from genotype by use of artificial neural networks. J Infect Dis 188, 653–660 (2003).
Larsen, P. E., Field, D. & Gilbert, J. A. Predicting bacterial community assemblages using an artificial neural network approach. Nat Methods 9, 621–625 (2012).
Beerenwinkel, N. et al. Geno2pheno: Estimating phenotypic drug resistance from HIV-1 genotypes. Nucleic Acids Res 31, 3850–3855 (2003).
Beerenwinkel, N. et al. Diversity and complexity of HIV-1 drug resistance: a bioinformatics approach to predicting phenotype from genotype. Proc Natl Acad Sci U S A 99, 8271–8276 (2002).
Xiang, Z. X. & Honig, B. Extending the accuracy limits of prediction for side-chain conformations. J Mol Biol 311, 421–430 (2001).
Xiang, Z., Soto, C. S. & Honig, B. Evaluating conformational free energies: the colony energy and its application to the problem of loop prediction. Proc Natl Acad Sci U S A 99, 7432–7437 (2002).
Case, D. A. et al. AMBER 12, University of California, San Francisco (2012).
Huang, G. B., Wang, D. H. & Lan, Y. Extreme learning machines: a survey. International Journal of Machine Learning and Cybernetics 2, 107–122 (2011).
Huang, G. B., Zhu, Q. Y. & Siew, C. K. Extreme learning machine: Theory and applications. Neurocomputing 70, 489–501 (2006).
Jakalian, A., Jack, D. B. & Bayly, C. I. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J Comput Chem 23, 1623–1641 (2002).
Ponder, J. W. & Richards, F. M. Tertiary Templates for Proteins - Use of Packing Criteria in the Enumeration of Allowed Sequences for Different Structural Classes. J Mol Biol 193, 775–791 (1987).
Soto, C. S., Fasnacht, M., Zhu, J., Forrest, L. & Honig, B. Loop modeling: Sampling, filtering and scoring. Proteins 70, 834–843 (2008).
Shenkin, P. S., Yarmush, D. L., Fine, R. M., Wang, H. J. & Levinthal, C. Predicting antibody hypervariable loop conformation. I. Ensembles of random conformations for ringlike structures. Biopolymers 26, 2053–2085 (1987).
Pettersen, E. F. et al. UCSF chimera - A visualization system for exploratory research and analysis. Journal of Computational Chemistry 25, 1605–1612 (2004).
Wang, Q. T. & Bryce, R. A. Improved Hydrogen Bonding at the NDDO-Type Semiempirical Quantum Mechanical/Molecular Mechanical Interface. J Chem Theory Comput 5, 2206–2211 (2009).
Palau, J., Argos, P. & Puigdomenech, P. Protein secondary structure. Studies on the limits of prediction accuracy. Int J Pept Protein Res 19, 394–401 (1982).
Cavasotto, C. N. & Phatak, S. S. Homology modeling in drug discovery: current trends and applications. Drug Discov Today 14, 676–683 (2009).
Vyas, V. K., Ukawala, R. D., Ghate, M. & Chintha, C. Homology Modeling a Fast Tool for Drug Discovery: Current Perspectives. Indian J Pharm Sci 74, 1–17 (2012).
Bolz, J., Farmer, I., Grinspun, E. & Schroder, P. Sparse matrix solvers on the GPU: Conjugate gradients and multigrid. Acm T Graphic 22, 917–924 (2003).
Kruger, J. & Westermann, R. Linear algebra operators for GPU implementation of numerical algorithms. Acm T Graphic 22, 908–916 (2003).
Owens, J. D. et al. GPU computing. P Ieee 96, 879–899 (2008).
Ryoo, S. et al. Program optimization carving for GPU computing. J Parallel Distr Com 68, 1389–1401 (2008).
Acknowledgements
This work is supported by the Hong Kong Research Grants Council (Project CityU 123809) and City University of Hong Kong (Project 7002843).
Author information
Authors and Affiliations
Contributions
V.L. carried out clinical observations on the patients, collected and provided the clinical data and participated in the design of the study. D.D.W. and W.Z. carried out the molecular structural studies and dynamics simulations of the proteins, participated in the design of the study, performed the statistical analysis and drafted the manuscript. H.Y. and M.W. initiated the project, participated in its design and coordination and helped draft the manuscript. All authors read and approved the final manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Table 1
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Wang, D., Zhou, W., Yan, H. et al. Personalized prediction of EGFR mutation-induced drug resistance in lung cancer. Sci Rep 3, 2855 (2013). https://doi.org/10.1038/srep02855
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep02855
This article is cited by
-
Prediction of sensitivity to gefitinib/erlotinib for EGFR mutations in NSCLC based on structural interaction fingerprints and multilinear principal component analysis
BMC Bioinformatics (2018)
-
Understanding the molecular basis of EGFR kinase domain/MIG-6 peptide recognition complex using computational analyses
BMC Bioinformatics (2015)
-
EGFR Mutant Structural Database: computationally predicted 3D structures and the corresponding binding free energies with gefitinib and erlotinib
BMC Bioinformatics (2015)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.






 ;
; .
.