Abstract
Folic acid (FA) has traditionally been associated with prevention of neural tube defects; more recent work suggests that it may also be involved in in the prevention of adult onset diseases. As the role of FA in human health and disease expands, it also becomes more critical to understand the mechanisms behind FA action. In this work we examined the hypothesis that folate receptor alpha (FRα) acts as a transcription factor. FRα is a GPI-anchored protein and a component of the caveolae fraction. The work described here shows that FRα translocates to the nucleus, where it binds to cis-regulatory elements at promoter regions of Fgfr4 and Hes1 and regulates their expression. The FRα recognition domain mapped to AT rich regions on the promoters. Until this time FRα has only been considered as a folate transporter, these studies describe a novel role for FRα as a transcription factor.
Similar content being viewed by others
Introduction
Traditionally folic acid (FA)1 has been associated with prevention of neural tube defects; however more recently FA has been associated with the prevention of adult onset disease, such as Alzheimer's disease, dementia, neuropsychiatric disorders, cardiovascular diseases and cerebral ischemia (reviewed in ref. 2). Cellular uptake of folate is mediated by specific carriers or receptors, including FRα (folate receptor alpha; also known as Folr1 and Folbp1)3, proton-coupled folic acid transporter (PCFT) and reduced folic acid carrier (RFC) (see ref. 4 for review). FRα, a GPI-anchored protein5 is critical for embryonic development6. Disruption of both FRα alleles in mice results in pups with a range of malformations and is lethal to the embryos at the time of neural tube closure6. FRα is one of the components of the caveolae fraction7, which includes EGFR8, caveolin-1 (Cav-1)9, the β subunit of heterotrimeric GTP binding protein (Gβ) and protein kinase C α subunit (PKCα)2. Cav-19 and EGFR10 act as transcription factors by binding to cis-regulatory elements of downstream target genes. EGFR binds to the promoters of cyclin D1, iNOS, B-Myb, Aurora-A, thymidylate synthase, COX-2, c-Myc and BCRP which are involved in tumorogenesis, chromosome instability and chemo-resistance11. Cav-1 binds to the promoters of cyclin D1 and FRα9, IGF-1 receptor12, BRCA113. This study examines a possible role of another caveolar protein, FRα as a transcription factor for key developmental genes.
Previous data from our lab2,14,15 demonstrated that FA remodels chromatin structures15. A second mechanism of FA action may be through FRα translocating to the nucleus and acting as a transcription factor. Bozard and colleagues16 reported the presence of FRα on the plasma membrane, the nuclear membrane and within endosomal structures; however the relevance of nuclear FRα was not determined. In this work we tested the hypothesis that in response to FA, FRα translocates to the nucleus and acts as a transcription factor.
To test the role of FRα as a transcription factor we examined nuclear localization in cell lines and interaction of FRα with two candidate genes FGFR4 and Hes1. These candidate genes were chosen because in our previous studies, working with neural stem cells from Pax3 mutant (also known as or Splotch (Sp−/−)) mouse embryos, we found that FA up-regulates Fgfr4 and Fgfr4 receptor protein2 and increases levels of Hes114.
Results
Nuclear localization of FRα
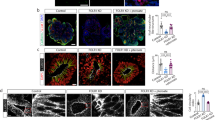
To test the hypothesis that FRα translocates to the nucleus, a time course (0 min, 15 min and 30 min) for FRα nuclear localization was performed in DAOY cells treated with FA. The results of FRα immunoblots using mouse monoclonal antibody on nuclear extracts (Fig. 1a, b) showed that FRα translocates to the nucleus within 15 min of FA incubation. It is to be noted that a very faint band of immunoreactivity for FRα (38 kd band) was present in the nucleus even in the absence of FA.
Nuclear localization of FRα.
(a) Nuclear extracts from DAOY cells treated with FA (200 μg/ml) for zero, 15 and 30 min at 37°C were subjected to immunoblotting using monoclonal anti-FRα (recognizing a 38 kd band) and polyclonal anti-pRB (recognizing 110 kd band) (see Supplementary Fig S1 ). (b) Ratio of FRα/pRB average band intensity (densitometry data is an average + SEM of three experiments). (c) Subcellular factions from DAOY cells not-treated or treated with FA (200 μg/ml) for 30 min were immunoblotted with NCAM, N-cadherin, ICAM-1 vimentin, hsp90, pRB, H3 and FRα (rabbit polyclonal) antibodies. The FRα polyclonal antibody is made against an epitope corresponding to amino acids 1-257 representing full length FR α of human origin. This antibody is reported to recognize multiple types of FR, α, β and perhaps γ. Rabbit IgG was used as a negative control. M, membrane enriched; C, cytosolic enriched; P, insoluble cytoskeletal enriched; N, nuclear enriched; CB, chromatin bound fraction. The data above is a representative example of 5 different western blots. (d) The data is the average of 5 different western blot experiments +/− standard error mean. The ratio of average band intensity of FRα (42 kd and 38 kd) to subcellular fraction markers: FRα/ICAM-1; FRα/hsp90; FRα/vimentin; FRα/pRB; FRα/H3 was determined using densitometry. Statistical significance was calculated using Student's t test. (e) The data in “d” is presented as total non-nuclear fraction comprising of membrane, cytosol and insoluble cytoskeletal pellet and total nuclear fraction comprised of nuclear and chromatin bound fractions. Statistical significance was calculated using Student's t test. (f) DAOY cells were grown in 8 well chamber slides in DMEM with 10% FBS for 24 h, then switched to serum free media in the absence or presence of FA (200 μg/ml) for 30 min at 37°C and immunostained using FRα monoclonal antibody and polyclonal pRB antibody and subjected to confocal microscopy. Secondary antibodies were donkey anti-rabbit Cy3 (red) and donkey anti-mouse Alexa488 (green). Yellow signals indicate co-localization of FRα (red) and pRB (green) in the nucleus (also stained blue with DAPI). The data is a representative of five separate experiments.
To study FRα distribution in the absence and presence of FA (30 min), we isolated different subcellular fractions of DAOY cells- membrane, cytosol, cytoskeletal, nuclear and chromatin enriched fractions and performed western immunoblots (Fig. 1c) using FRα antibody along with antibodies against subcellular markers NCAM, N-cadherin and I-CAM1 (for membrane enriched fraction), hsp90 (for cytosolic enriched fraction), vimentin (for cytoskeletal enriched fraction), pRB (for nuclear enriched fraction) and histone H3 (for chromatin bound fraction). The ratio of the average band intensities of the two immunoreactive bands of FRα (42 kd and 38 kd doublet) with the marker of individual subcellular fraction (FRα/ICAM-1, FRα/hsp90, FRα/vimentin, FRα/pRB and FRα/H3 bands) were determined using densitometry (Fig. 1d). It is to be noted that all the membrane markers used here also showed strong immunoreactivity in the nuclear enriched fraction. The hsp90 immunoreactivity was highest in the cytosolic enriched fractions (C) and the vimentin antibody cross-reacted with the insoluble cytoskeletal pellet (P). The pRB immunoreactivity was highest in the nuclear enriched fractions (N) and histone H3 antibody immunoreacted with the chromatin bound fraction (CB). In the absence of FA, FRα was predominantly present in the cytosolic fraction whereas in the presence of FA, the FRα (42 kd band) appeared to translocate to the non-nuclear fraction (membrane and cytoskeletal pellet fraction) and the 38 kd band to the nucleus. In the nucleus the FRα (38 kd band) appeared to be present in the chromatin bound fraction in the presence of FA. When the data in Fig. 1e is presented as total non-nuclear fraction (membrane + cytosol + insoluble cytoskeletal pellet) and nuclear fraction (nuclear + chromatin bound) we observe that even in the absence of FA, FRα is present in the nucleus and in the presence of FA, there is a significant increase in the translocation of FRα to the nuclear fraction.
Presence of FRα in the nucleus was further confirmed by confocal microscopy in DAOY cells (Fig. 1f). pRB was chosen as a nuclear marker. Increased co-localization of FRα and pRB was observed in the presence of FA. These results suggest the following: (i) In the absence of FA, there is a more FRα in the cytosolic fraction; (ii) Upon FA treatment, FRα is distributed significantly to the non-nuclear membrane fraction as well to the nuclear enriched and chromatin bound fractions; (iii) Of the two immunostained FRα- 42 kd and 38 kd bands, the 42 kd band seems to translocate to the membrane enriched fraction in the presence of FA. Although both 42 kd and 38 kd bands of FRα appear to translocate to the nucleus, only the 38 kd band translocates significantly to the chromatin bound fraction in the FA treated cells.
FRα binds to cis-regulatory elements of gene promoters
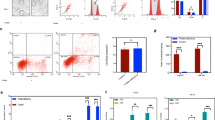
The above studies suggested that FRα translocates to the nucleus and in the presence of FA, it is enriched in the chromatin bound fraction. To determine whether FRα activates FGFR4, FGFR4 promoter-luciferase constructs P-535/+99 from human FGFR4 promoter17 were transiently transfected into DAOY cells, treated or not treated with FA. FGFR4 promoter-luciferase reporter activity increased (p<0.05) in the presence of FA (Fig. 2a). FRα significantly increased FGFR4 promoter-luciferase in the absence of FA (p<0.001), with a further increase in the presence of FA (p<0.0001). This demonstrates that FRα activates FGFR4 promoter by binding to cis-regulatory elements.
Activation of Pax3 downstream target genes by FRα.
FRα-pcDNA3 or control pcDNA3 (0.2 ng/well) constructs were co-transfected with Hes1 promoter-luciferase construct (a), human FGFR4 promoter-luciferase construct P-535/+9919 (b) or a control PGL3 (promoter-less luciferase gene) into DAOY cells.FA (200 μg/ml) was added 24 h post transfection. Luciferase activity was assayed 48 h post-transfection. pRLnull (5 ng/well) was used as a transfection control in all wells. For both Hes1 promoter luciferase and FGFR4 promoter-luciferase construct P-535/+99 FRα significantly increased promoter activity, with the highest increases observed in the presence of FA. Experiments were performed in quadruplicate with each data point in duplicate. * p<0.05; ** p<0.001; *** p<0.0001 (Student T-test).
To establish if FRα activates other FA modulated genes by binding to cis-regulatory regions, mouse Hes1 promoter-luciferase construct18 was co-transfected with FRα expression vector into DAOY cells treated or not treated with FA. Hes1 is a Pax3 downstream target gene18, FA increases Hes1 mRNA and protein levels14. Hes1-promoter-luciferase reporter activity increased (p<0.05) in response to FA. FRα significantly increased (p<0.001) Hes1-promoter-luciferase without FA (Fig. 2b), with a further increase with FA treatment. These data indicate that FRα transcriptional activation is not limited to FGFR4.
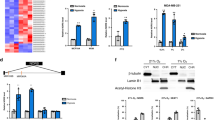
To confirm FRα binding to cis-regulatory elements of Hes1 and Fgfr4 promoters in intact embryos, chromatin immunoprecipitation (ChIP) experiments were performed using the lower lumbar region of the neural tube from wild-type (WT) mouse embryos (E10.0, 30 somite stage), an area where both of these genes are expressed. FRα bound to cis-regulatory regions of Hes1 and Fgfr4 promoters in vivo (Fig. 3a).
FRα binds to murine Hes1 and Fgfr4 promoter cis-regulatory elements.
(a) ChIP assays was performed using E10.0 (30 somite) lumbar neural tube. Anti-FRα polyclonal antibody was used to immunoprecipitate (IP) the protein–DNA complex. This antibody is made against epitope corresponding to amino acids 1-257 representing full length FR α of human origin. This antibody is reported to recognize multiple types of FR, α, β and perhaps γ. Primers used to amplify cis-regulatory elements in Hes1 and Fgfr4 promoters are shown in Supplementary Information Table 1. Rabbit IgG was used as an IP negative control. ChIP experiments were performed in triplicate using one lumbar neural tube region per assay with a total of n = 4. (b) EMSA of binding reactions performed using GST-FRα fusion protein and 32P-labeled double-stranded oligonucleotides. Mouse Hes1 oligo #1 (with AANTT): 5′-AAAAAATTATTTTTTTTTTGCGTGAAG-3′; Mouse Hes1 oligo #2 (mutant AAA>CCC): 5′-AAACCCTATTTCCCCTTTGCGTGAAG-3′: (c)Mouse Fgfr4 oligo #3 (with AANTT): 5′-CAAACAAACAAAAAGAAACAACAAAAAAACTTTTTA-3′; Mouse Fgfr4 oligo #4 (with AANTT): 5′-ATAAAAGCACAACTTTTTACAAAGTTTAAAGTTTTTT-3; Mouse Fgfr4 oligo #5 (deletion mutant without AANTT) 5′-CGTTCGCGTGCAGTCCGAGATAT-3′. The arrow shows GST-FRα binding to oligonucleotides which have the AANTT sequence.
FRα binds to AANTT consensus sequence on Hes1 or FGFR4 promoter
To identify putative FRα binding sequences in Hes1 and Fgfr4 promoters, 32P-labeled oligonucleotides were made from appropriate promoter regions and EMSA was performed using affinity-purified GST-FRα fusion protein (Fig. 3b, c). GST-FRα fusion protein bound the Hes1 oligonucleotide 5′-AAAATTATTTTTTTTTTGCGTGAAG-3′ which had AANTT or NAAAAN and/or NTTTTN sequences. When this sequence was mutated as in 5′-AACCCTTATTCCCCTTTGCGTGAAG-3′ there was no shift. Similarly, on the Fgfr4 promoter the GST-FRα binding site mapped to AANTT or NAAAAN and/or NTTTTN in the oligonucleotide 5′-CAAACAAACAAAAAGAAACAACAAAAAAACTTTTTA-3′ and NTTTTTN in the oligonucleotide 5′-ATAAAAGCACAACTTTTTACAAAGTTTAAAGTTTTTT-3′. When the oligonucleotide sequence did not have the AANTT or TTNAA and NAAAAN consensus GST-FRα did not show a shift as in the case of 5′-CGTTCGCGTGCAGTCCGAGATAT-3′.
To further confirm the identity of FRα binding sites on Hes1 and FGFR4 promoters, AANTT sites were mutated on Hes1 and FGFR4 promoter-luciferase reporter constructs P-535/+99. Mutated constructs were transfected into DAOY cells as above. Luciferase activity did not increase with these constructs for either Hes1 or FGFR4 promoters (Fig. 4a). The results confirm that FRα binds Hes1 and FGFR4 promoters at AANTT or TTNAA and NTTTTN or NAAAAN sites.
FA does not activate Hes1 or FGFR4 promoter luciferase activity when the FRα consensus sequence (AANTT) is mutated.
(a) AA>CC substitution mutations were made at the putative FRα binding sites on Hes1 or FGFR4 promoters (mutated sites are shown in the supplemental information). Hes1 promoter-luciferase containing plasmid or plasmids containing mutated sequences (20 ng) were transiently co-transfected with FRα- pcDNA3 or pcDNA3 vector control into DAOY cells treated or not treated with FA and luciferase assays were performed. Renilla luciferase plasmid, pRLnull (5 ng/well) was simultaneously transfected as an insertional control for transfection efficiency. FRα-pcDNA3 values were expressed as the activity of Firefly-luciferase, minus values obtained for control pcDNA3 transfection. Experiments were performed in triplicate with each data point in duplicate; *p<0.05; **p<0.001; ***p<0.0001 (Student T-test). (b) A hypothetical working model depicting FRα as a transcription factor. FRα, a GPI-anchored protein, gets internalized in a caveolar structured early endosomes, which undergo acidification and subsequent fusion with lysosomes. GPI-specific phospholipase D cleaves FRα from its GPI-anchor. FRα is released and translocates to the nucleus via an unknown mechanism(s) where it binds cis-regulatory elements of different gene promoters.
Discussion
Previous work from our lab demonstrated that in the absence of functional Pax3, FA increased KDM6B, through up-regulation of KDM6B targeting micro-RNAs. This in turn altered H3K27 methylation marks on the promoters of Pax3 downstream targets, Hes1 and Neurog2 and affected gene transcription14. Thus one mechanism of FA action is through remodeling of chromatin structures. In this study we have elucidated a second mechanism for FA action, through activation of FRα and its' subsequent action as a transcription factor. A hypothetical model showing FRα internalization is presented in Fig. 4b. FRα is internalized in a caveolar structure as early endosome19. The endosome becomes increasingly acidic20 and fuses with a lysosome21. In the lysosome FA is released22 and lysosomal GPI-specific phospholipase D23 cleaves off the GPI anchor on FRα, which is then set free. Free FRα translocates into the nucleus where it binds to cis-regulatory elements of target genes and directly activates transcription. This model does not take into account FRα recycling and it is still unclear exactly how FRα translocates into the nucleus.
Binding to Hes1 and Fgfr4 promoters suggests FRα involvement in stem cell maintenance and skeletal muscle differentiation, respectively. The list of putative FRα targets shown in Table 1, suggests that FRα may be involved in regulating a plethora of developmental genes involved in myogenesis, skeletonogenesis, cell mobility, neural crest cell migration, cranial and cardiac neural crest formation, hair morphogenesis, oligodendrogenesis, spermatogenesis24, melanogenesis25 and epithelial to mesenchymal transformation26. A survey of the promoter regions of c-Met, PDGFα, TGFβ2, MITF, N-CAM, c-RET, MyoD and Tyrp-1 indicates that the FRα binding motif AANTT or NTTTTN and/or NAAAAN map close to Pax3, a transcription factor and multifunctional regulatory protein, expressed early in embryogenesis, binding sites, which map to ATTA, GTTCC, TAAT, CCTTG, CAAGG, GTTAT, TATTG, GTGTGA and CAGTGT27. These observations suggest that FRα and Pax3 might appear as a complex regulating target genes synergistically. However, up-regulation of Hes1 or Neurog2 in FA-rescued Sp−/− embryos14 suggests that FRα may also have a role independent of Pax3. Future work shall test this hypothesis.
The observation that FRα acts like a transcription factor is relevant to our understanding of the mechanisms of FA action during development and has significant implications for disorders associated with FA deficiency and FRα misregulation and for management of human cancers which express FRα as a tumor antigen. FA has been associated with the prevention of adult onset diseases (reviewed in ref. 2). In some of these cases FA deficiency may not be the problem. The issue may be an inability to respond to FA due to misregulation of FRα. Cazzaniga and colleagues28 compared levels of serum folate and assessed differences in folate binding ability with primary fibroblast cultures, from Alzheimer's disease (AD) patients and age-matched healthy subject. Circulating folate was significantly lower in AD patients, whereas folate binding to fibroblasts was significantly higher, possibly due to enhanced expression of FRα in AD fibroblasts. Cerebral folate transport deficiency is an inherited brain-specific folate transport defect caused by mutations in the folate receptor 1 (FOLR1) gene which codes for FRα29. This disorder generally has a late infantile onset and symptoms include progressive movement disturbance, psychomotor decline, epilepsy and disturbed brain myelination, as well as a depletion of white matter choline and inositol30. Grapp and colleagues29 reported that whereas WT FRα was localized in the plasma membrane, in cerebral folate deficiency FRα mutants were mistargeted to intracellular compartments. The data presented in this paper provides relevant insight to these clinical situations. If FA interaction with FRα is misregulated, key transcriptional events may be affected. This in turn can lead to a series of developmental consequences or to adult onset disease associated with FA levels. Further work needs to be done to examine direct transcriptional activation of FRα responsive genes by FRα and its' role in these multifactorial diseases.
Another clinical role for FRα is in cancer, where it is recognized as a tumor antigen/biomarker31. Because of this, diagnostic and therapeutic methods which exploit FRα are being developed for cancer treatment, including the use of folate-drug conjugates31. The knowledge that FRα acts as a transcription factor can be exploited to target FA-siRNA or FA-drug conjugates to silence downstream targets in appropriate cancers. For instance, two Pax3 downstream targets c-MET and MITF are associated with melanoma32. MET promotes the melanoma phenotype by stimulating migration, invasion, resistance to apoptosis and tumor cell growth. PAX3 mediates MET induction through direct activation of the gene and indirect regulation through MITF. FA-drug conjugates exploiting the proximity of Pax3 and FRα binding sites could potentially silence c-met and/or MITF expression.
In summary, our study shows that FRα is localized in the nucleus, where it binds to cis-regulatory elements (AANTT or TTNAA and NTTTTN or NAAAAN) on FA modulated genes and activates their transcription. This novel role of FRα as a transcription factor provides insight into developmental mechanisms associated with FA responsiveness. It also provides an exciting new avenue to explore for treatment of diseases associated with FA deficiency, FRα misregulation and cancers which express FRα as a biomarker.
Methods
Antibodies and reagents
pRb antibody-rabbit polyclonal; 1:1000, Cell Signaling Technologies: Ser807/811, FRα antibody-rabbit polyclonal; 1:500. Santa Cruz: sc-28997), NCAM (Santa Cruz; sc-1507), N-Cadherin (Santa Cruz; sc-1502), I-CAM-1(Santa Cruz; sc-1510), vimentin (BD Pharmingen; 550513); pRB (Cell Signaling Technologies, Ser807/811 rabbit polyclonal; 1:1000); Histone H3 (Cell Signaling Technologies, 9701, rabbit polyclonal 1:1000); Mouse monoclonal anti-FRα antibody from Lifespan biosciences (cat # LS-C23683). Donkey anti-rabbit IgG-HRP (sc-2305) and Donkey anti-mouse IgG-HRP (sc-2306) were from Santa Cruz. DAPI was purchased from Sigma. Secondary antibodies for immunostaining procedures were donkey anti-rabbit Cy3 (red) (1:200) and donkey anti-mouse Alexa488 (green) (1:200).Primers and oligonucleotides for EMSA were from Operon. Wild type (WT) C57BL/6J male and female mice were from Jackson Labs. For timed embryos, females and males were mated, the morning a vaginal plug was observed was noted as E0.5. Pregnant dams were euthanized by cervical dislocation with CO2 inhalation and moniliform uterine beads were removed at E10.5. Neural tubes were dissected out as described earlier33. All animal experiments were approved by IACUC -Children's Hospital of Chicago Research Center, Chicago (Approval # Mayanil, IACUC ID: 13-001.0 09) and all experiments were performed in accordance with relevant institutional guidelines and regulations.
Statistical analysis
Values given are means: SEM. Probabilities (p) were calculated with Student's unpaired t test using GraphPad Prism version 4.0. p values < 0.05 were considered statistically significant. One-way ANOVA with Bonferroni's multiple comparison tests were used for multiple comparisons between data.
Nuclear localization of FRα
DAOY cells were treated with FA (200 μg/ml) for zero, 15 and 30 minutes at 37°C. For the western blots studies, the DAOY cells not-treated or treated with FA for 30 min were used. Subcellular fractions, membrane enriched, cytosolic, insoluble pellet cytoskeletal fraction, nuclear and chromatin bound fractions (30 μg) were immunoblotted with antibodies against FRα (rabbit polyclonal, 1:500), NCAM (1:500); N-Cadherin (1:1000); I-CAM-1 (1:1000); vimentin (1:10,000); pRB (1:500) and Histone H3 (1:500). This rabbit polyclonal anti-FRα antibody is made against epitope corresponding to amino acids 1-257 representing full length FR α of human origin. This antibody is reported to recognize multiple types of FR, α, β and perhaps γ. The average band intensity of FRα/ICAM-1; FRα/hsp90; FRα/vimentin; FRα/pRB; FRα/H3 was determined using densitometry. For immunostaining, DAOY cells were plated and grown in DMEM with 10% FBS for 24 hours and then changed to serum-free media. FA (200 μg/ml) was added to appropriate wells. The cells were allowed to grow for an additional 30 min. Cells were immunostained with anti-FRα (mouse monoclonal antibody; 1:100) and pRB (rabbit polyclonal; 1:100). This mouse monoclonal FRα antibody recognizes only the 38 kd band of FRα. Rabbit IgG was used as a negative control. Secondary antibodies were donkey anti-rabbit Cy3 (red) (1:200) and donkey anti-mouse Alexa488 (green) (1:200). Confocal microscopy was done with a Zeiss 510 META Confocal Laser Scanning Microscope.
Real time quantitative RT-PCR
Real time quantitative RT-PCR was done as described earlier27. Primers and probes used in this study were designed using Primer Express software (PerkinElmer Life Sciences). Primers were synthesized by Operon Inc. probes were synthesized by MegaBases Inc (Refer Supplementary Information-Table 1 for primers).
Chromatin immunoprecipitation (ChIP) assays
ChIP assays using lumbar neural tube from WT embryos (E10.0) were performed as described earlier34. PCR was performed with primers for murine Hes1 and Fgfr4 promoter regions (Supplementary Information-Table S1 for primers). All ChIP samples were tested for false-positive PCR amplification by sequencing the 200-bp amplified product to ascertain the specificity of FRα binding to cis-regulatory elements.
Analysis of Hes1 and Fgfr4 promoter activity
A 2.5 kb Hes1 promoter-luciferase construct was provided by Dr. R. Kageyama, Institute for Virus Research, Kyoto University Kyoto, Japan. Human FGFR4 promoter constructs were provided by Dr Shereen Ezzat, Departments of Medicine, Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Canada. The FRα expression construct in pcDNA3 was kindly provided by Dr Asok Antony, Indiana University Medical School Indianapolis, IN, USA. Renilla luciferase plasmid, pRL-null (0.5 μg) (dual luciferase system, Promega), was simultaneously transfected as an insertional control for transfection efficiency in all of the studies. DAOY cells were seeded at 5 × 104 cells/60 mm diameter dish in DMEM supplemented with 10% fetal calf serum for 24 h prior to transfection. For co-transfections: human FRα cDNA and mouse Hes1-Luc promoter or human FRα cDNA and human FGFR4-Luc promoter were transfected into the cells using MegaTran 1.0 from OriGene; Cat#: TT200003. After 24 h, FA was added to the treatment wells. The cells were washed 3 times with phosphate-buffered saline (PBS) and lysed with Passive Lysis Buffer (PLB) 48 h post transfection. Luciferase activity was measured using a Vector 2 Luminometer (PerkinElmer Life Sciences). The AA>CC substitution mutations of the putative FRα binding sites on murine Hes1 or human FGFR4 promoters were made with the QuickChangeXL site-directed mutagenesis kit (Stratagene). Hes1 promoter-luciferase containing plasmid or plasmids containing the mutated sequences (20 ng) were transiently co-transfected with FRα- pcDNA3 or pcDNA3 vector control into DAOY cells and luciferase assays were done as described earlier34 using the Dual Luciferase kit from Promega. Renilla luciferase plasmid, pRL-null (5 ng/well) (Dual Luciferase System Promega), was simultaneously transfected as an insertional control for transfection efficiency.
Purification of GST-FRα fusion protein
GST-FRα fusion plasmid was kindly provided by Dr. Asok Antony. Escherichia coli was transformed with GST-FRα fusion plasmid and the cells were grown overnight in LB medium. GST-FRα fusion protein production was induced with 0.1 mM isopropyl-D-thiogalactopyranoside (Sigma) for 2 hr. Cells were pelleted and sonicated to release the protein in 50 mM Tris-HCl pH7.8 buffer. Supernatant containing GST-FRα fusion protein among other proteins was loaded onto glutathione-Sepharose 4B column (Amersham Pharmacia Biotech). Unbound proteins were washed with 50 mM Tris-HCl pH7.8 buffer and eluted with 10 mM reduced glutathione in 50 mM Tris-HCl, pH 9.5. Eluted protein was concentrated using Centricon 10 (Amicon) as per the manufacturer's instruction.
Electro-mobility shift assays (EMSA)
Probes were prepared for EMSA by annealing complementary oligonucleotides representing selected regions of murine Hes1 and Fgfr4 promoters, followed by 5′-end labeling with [γ-32P] ATP by T4 polynucleotide kinase. EMSA was done as described earlier18. Double-stranded oligonucleotide probes produced, covered the following regions: WT Hes1 promoter oligo: 5′-AAAAAATTATTTTTTTTTTGCGTGAAG-3′ and mutant Hes1 promoter oligo: 5′-AAACCCTTATTCCCCTTTTGCGTGAAG-3′; WT Fgfr4 promoter oligo #1: 5′-CAAACAAACAAAAAGAAACAACAAAAAAACTTTTTA-3′ WT promoter oligo #2: 5′-ATAAAAGCACAACTTTTTACAAAGTTTAAAGTTTTTT-3′ and an oligo without AANTT sequence #3: 5′-CGTTCGCGTGCAGTCCGAGATAT-3′. For shift assays 4 μg GST-FRα fusion protein was pre-incubated at room temperature for 30 min with 4 M urea and EMSA reaction buffer (100 mM Tris-HCl PH7.5, 500 mM KCl, 6.5% glycerol, 50 mM pyrophosphate, 25 mM DTT in 2.5% Tween 20, 1 μg/μl salmon sperm DNA) prior to addition of labeled oligonucleotides. For hot reactions GST-FRα fusion protein and radio-labeled oligonucleotides were added and pre-incubated for 30 min at room temperature. For cold reactions GST-FRα fusion protein and unlabeled oligonucleotides were added and labeled oligonucleotides were added after 30 min. After pre-incubation, free DNA and DNA protein complexes were resolved in 6% polyacrylamide gels (these gels were pre-run at 1000 V and 5 mA for 16 h in the continuous cooling system) using 0.25× TBE as the running buffer. Electrophoresis was performed at 1000 V and 25 mA for 2 h in a continuous cooling system. To visualize shifted bands, gels were dried at RT and transferred to Phosphor Imager Screens (Amersham Biosciences). Gels were exposed overnight at 4°C.
References
Wlodarczyk, B. J., Tang, L. S., Triplett, A., Aleman, F. & Finnell, R. H. Spontaneous neural tube defects in splotch mice supplemented with selected micronutrients. Toxicol. Appl. Pharmacol. 213, 55–63 (2005).
Mayanil, C. S. et al. Maternal intake of folic acid and neural crest stem cells. Vitam. Horm. 87, 143–173 (2011).
Zhu, H. et al. Differentially expressed genes in embryonic cardiac tissues of mice lacking Folr1 gene activity. BMC Dev Biol. 7, 128 (2007).
Zhao, R., Diop-Bove, N., Visentin, M. & Goldman, I. D. Mechanisms of membrane transport of folates into cells and across epithelia. Annu. Rev. Nutr. 31, 177–201 (2011).
Ratnam, M., Marquardt, H., Duhring, J. L. & Freisheim, J. H. Homologous membrane folate binding proteins in human placenta: Cloning and sequence of a cDNA. Biochemistry 28, 8249–8254 (1989).
Piedrahita, J. A. et al. Mice lacking the folic acid-binding protein Folbp1 are defective in early embryonic development. Nat. Genet. 23, 228–232 (1999).
Smart, E. J., Ying, Y. S., Mineo, C. & Anderson, R. G. A detergent-free method for purifying caveolae membrane from tissue culture cells. Proc. Natl. Acad. Sci. U S A. 92, 10104–10108 (1995).
Goldstein, J. L., Brown, M. S., Anderson, R. G. W., Russell, D. W. & Schneider, W. J. Annu. Rev. Cell Biol. 1, 1–39 (1985).
Sanna, E. et al. Binding of nuclear caveolin-1 to promoter elements of growth-associated genes in ovarian carcinoma cells. Exp. Cell Res. 313, 1307–1317 (2007).
Lin, S. Y. et al. Nuclear localization of EGF receptor and its potential new role as a transcription factor. Nat. Cell Biol. 3, 802–808 (2001).
Wang, Y. N. & Hung, M. C. Nuclear functions and subcellular trafficking mechanisms of the epidermal growth factor receptor family. Cell. Biosci. 2, 13 (2012).
Glait, C. et al. Caveolin-1 up-regulates IGF-I receptor gene transcription in breast cancer cells via Sp1- and p53-dependent pathways. Exp. Cell Res. 312, 3899–3908 (2006).
Glait, C., Ravid, D., Lee, S. W., Liscovitch, M. & Werner H. Caveolin-1 controls BRCA1 gene expression and cellular localization in human breast cancer cells. FEBS Lett. 580, 5268–5274 (2006).
Ichi, S. et al. Folic acid remodels chromatin on Hes1 and Neurog2 promoters during caudal neural tube development. J. Biol. Chem. 285, 36922–36932 (2010).
Ichi, S. et al. Fetal neural tube stem cells from Pax3 mutant mice proliferate, differentiate and form synaptic connections when stimulated with folic acid. Stem Cells Dev. 21, 321–330 (2012).
Bozard, B. R. et al. Molecular and biochemical characterization of folate transport proteins in retinal Müller cells. Invest. Ophthalmol. Vis. Sci. 51, 3226–3235 (2010).
Ezzat, S., Yu, S. & Asa, S. L. Ikaros isoforms in human pituitary tumors: distinct localization, histone acetylation and activation of the 5' fibroblast growth factor receptor-4 promoter. Am. J. Pathol. 163, 1177–1184 (2003).
Nakazaki, H. et al. Key basic helix-loop-helix transcription factor genes Hes1 and Ngn2 are regulated by Pax3 during mouse embryonic development. Dev. Biol. 316, 510–523 (2008).
Turek, J. J., Leamon, C. P. & Low, P. S. Endocytosis of folate-protein conjugates: ultrastructural localization in KB cells. J. Cell. Sci. 106, 423–430 (1993).
Lee, R. J., Wang, S. & Low, P. S. Measurement of endosome pH following folate receptor-mediated endocytosis. Biochim Biophys Acta. 1312, 237–242 (1996).
Luzio, J. P., Gray, S. R. & Bright, N. A. Endosome-lysosome fusion. Biochem. Soc. Trans. 38, 1413–1416 (2010).
Sabharanjak, S. & Mayor, S. Folate receptor endocytosis and trafficking. Adv. Drug Deliv. Rev. 56, 1099–1109 (2004).
Hari, T., Kunze, H., Bohn, E., Brodbeck, U. & Bütikofer, P. Subcellular distribution of glycosylphosphatidylinositol-specific phospholipase D in rat liver. Biochem. J. 320, 315–319 (1996).
Andrae, J., Gallini, R. & Betsholtz, C. Role of platelet-derived growth factors in physiology and medicine. Genes Dev. 22, 1276–1312 (2008).
Widlund, H. R. & Fisher, D. E. Microphthalamia-associated transcription factor: a critical regulator of pigment cell development and survival. Oncogene 22, 3035–3041 (2003).
Huang, S. S. & Huang, J. S. TGF-beta control of cell proliferation. J. Cell. Biochem. 96, 447–462 (2005).
Mayanil, C. S. et al. Microarray analysis detects novel Pax3 downstream target genes. J. Biol. Chem. 276, 49299–49309 (2001).
Cazzaniga, E., Bulbarelli, A., Lonati, E., Re, F., Galimberti, G. et al. Enhanced folate binding of cultured fibroblasts from Alzheimer's disease patients. Neurosci. Lett. 436, 317–320 (2008).
Grapp, M. et al. Molecular characterization of folate receptor 1 mutations delineates cerebral folate transport deficiency. Brain 135, 2022–2031 (2012).
Dill, P. et al. Pyridoxal phosphate-responsive seizures in a patient with cerebral folate deficiency (CFD) and congenital deafness with labyrinthine aplasia, microtia and microdontia (LAMM). Mol. Genet. Metab. 104, 362–368 (2011).
Vlahov, I. R. & Leamon, C. P. Engineering Folate-Drug Conjugates to Target Cancer: from Chemistry to Clinic. Bioconjug. Chem. 23, 1357–1369 (2012)
Mascarenhas, J. B. et al. PAX3 and SOX10 activate MET receptor expression in melanoma. Pigment Cell Melanoma Res. 23, 225–237 (2010).
Ichi, S. et al. Role of Pax3 acetylation in the regulation of Hes1 and Neurog2. Mol. Biol. Cell 22, 503–512 (2011).
Mayanil, C. S. et al. Regulation of murine TGFβ2 by Pax3 during early embryonic development. J. Biol. Chem. 281, 24544–24552 (2006).
Acknowledgements
This work was supported by the State of Illinois Excellence in Academic Medicine award (C. S. M.), a Grant from the Spastic Paralysis Research Foundation of Illinois-Eastern Iowa District of Kiwanis (C. S. M. and D. G. M.), the Spina Bifida Association and CHCRC Pilot Grant award (C. S. M.). We thank Dr. R. Kageyama, Institute for Virus Research, Kyoto University Kyoto, Japan, for providing the Hes1 promoter-luciferase construct; Dr Shereen Ezzat, Departments of Medicine, Laboratory Medicine and Pathobiology, University of Toronto, Toronto, Canada for providing Human FGFR4 promoter constructs; Dr Asok Antony, Indiana University Medical School, Indianapolis, IN, USA for providing the FRα expression construct in pcDNA3 and GST-FRα plasmids; and William Goossens, manager of the Microscopy and Imaging Facility at Children's Hospital of Chicago Research Center for assistance with confocal microscopy. We thank Bio-Rad for their generous gift of Trans-Blot Turbo and the ChemiDocMP system.
Author information
Authors and Affiliations
Contributions
V.B., K.S., C.S.M. designed research; V.B., E.S. and S.I. performed confocal microscopy, V.B., K.S. and T.T. performed EMSA and luciferase assays. V.B., X.I. and B.M.F. made promoter luciferase mutants. K.S. and T.T. purified GST-FRα fusion protein. V.B., G.X. and S.I. did statistical analysis. T.T., D.G.M. and C.S.M. coordinated the work. B.M.F. and C.S.M. interpreted the data and C.S.M. wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Supplementary Information
Supplementary Information
Rights and permissions
This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivs 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Boshnjaku, V., Shim, KW., Tsurubuchi, T. et al. Nuclear localization of folate receptor alpha: a new role as a transcription factor. Sci Rep 2, 980 (2012). https://doi.org/10.1038/srep00980
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep00980
This article is cited by
-
microRNA-622 upregulates cell cycle process by targeting FOLR2 to promote CRC proliferation
BMC Cancer (2024)
-
Folate receptor overexpression induces toxicity in a diet-dependent manner in C. elegans
Scientific Reports (2024)
-
Noncanonical function of folate through folate receptor 1 during neural tube formation
Nature Communications (2024)
-
Excretable, ultrasmall hexagonal NaGdF4:Yb50% nanoparticles for bimodal imaging and radiosensitization
Cancer Nanotechnology (2021)
-
Reinforcing one-carbon metabolism via folic acid/Folr1 promotes β-cell differentiation
Nature Communications (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.