« Prev Next »
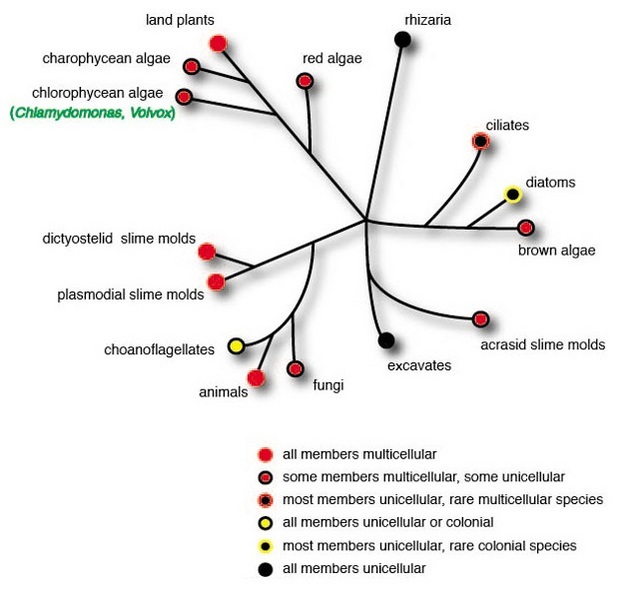
Life is very good at reinventing itself over time, and one of its most important innovations has been multicellularity, the capacity to make multiple cells and cell types that carry out specialized functions. Without the evolution of multicellularity, our planet would be a very different place — a world without plants or animals of any kind, and of course without humans. Yet even though multicellular species have evolved independently in most major lineages of eukaryotic organisms — including not only those to which plants and animals belong, but also green algae, brown algae, red algae, ciliates, slime molds, and fungi — we know surprisingly little about how this evolution came about (Figure 1). Do certain properties predispose a unicellular lineage to make the leap to multicellularity? Are certain types of genes/gene families, or genetic mechanisms especially important for this sort of transition to occur? Does the evolution of multicellularity require big steps involving major increases in genome size and/or expansions in gene families, or even many new kinds of genes? Or might the transition to a multicellular form possibly take place in smaller steps, involving only subtle changes? Scientists who study a family of green algae that includes unicellular Chlamydomonas and multicellular Volvox are beginning to find answers to some of these questions.
What Is Multicellularity?
Before we delve into these questions, note that not all multicellular lifestyles are the same. Many species of multicellular organisms differ greatly from each other with respect to the types of developmental mechanisms and traits they have evolved. For instance, by definition every multicellular organism possesses multiple cells that remain associated following cell division. But while plant and animal species generate at least a dozen different types of cells, with groups of cells organized into tissues and/or organs, some multicellular organisms, such as slime molds and at least one ciliate species, possess very few cell types and do not produce tissues or organs that are themselves composed of multiple cell types. Furthermore, animal embryos undergo gastrulation, a process by which groups of cells migrate or change position with respect to each other, but plant embryos do not. It turns out there are many different ways to be multicellular.
Good Model Systems for Multicellularity Are Rare
There is a very simple reason we know so little about how multicellularity arises: The phenomenon is very difficult to investigate. One complicating factor is that most transitions to multicellularity, such as the ones that gave rise to plants and animals, occurred deep in the past, approaching a billion years ago (Sanderson 2003; Peterson & Butterfield 2005). Why does it matter how long ago multicellularity evolved? The longer ago divergence from a common unicellular ancestor of existing multicellular and unicellular species occurred, the more genetic changes have accumulated that are irrelevant to multicellularity. These accumulated changes make it harder to sift though and determine exactly which genetic changes account for the transition to multicellularity.
Another thing that makes evolution of multicellularity difficult to study is the challenge in finding a good set of existing organisms to compare — a good model system. Few unicellular-multicellular species sets are suitable for this type of comparison, as a true system for experimental testing, because the two sets must be closely related. The reason why the best-known animal model systems (fruit flies, nematodes, zebrafish, mice, etc.) and plant model systems (Arabidopsis, corn, rice, tobacco, etc.) are not well suited is that their closest unicellular relatives — the choanoflagellates (animals) and charophycean algae (plants) — diverged from the multicellular lineage too long ago, so they have relatively few genetic similarities to animals and plants.
An Ideal Model System: Volvox, Chlamydomonas, and the Volvocine Green Algae
Cells of one type, called somatic cells, number about 2,000 and closely resemble Chlamydomonas unicells. Somatic cells are small, have two flagella, and reside in a monolayer at the surface of a sphere of gelatinous extracellular matrix (ECM). Their job is to swim and keep Volvox in the light so that it can photosynthesize. Unlike Chlamydomonas unicells, Volvox somatic cells cannot divide, and this distinction is very important — Volvox has multicellularity with division of labor because its somatic cells lost the capacity for reproduction. Reproduction is carried out by a second type of specialized cell, called the gonidium. Gonidia are large and do not have flagella (see Figure 2E), so they cannot swim (and must therefore rely on somatic cells for motility), but they can divide. Each of the approximately sixteen gonidia has the capacity to generate a new individual through a series of ten to eleven embryonic cell divisions that generate all the cells present in the next generation. Is this sort of division of labor unique to Volvox? Probably not. Some scientists believe that the segregation of somatic functions (like swimming) and reproduction into distinct cell types was one of the first key steps in the evolution of multicellularity in animals as well (Buss 1987; King 2004).
Researchers believe that the last common ancestor of the present-day volvocine algae was a unicellular species closely resembling modern-day Chlamydomonas and that Chlamydomonas may not have changed much at the genetic level, with respect to that ancestor (Kirk 2005; Herron & Michod 2008). Researchers also know, on the basis of information from plant and algal fossils and from molecular clock analyses that members of the volvocine family have been diverging from each other for only about 200 million years (Herron et al. 2009). Combined with the fact that both Volvox and Chlamydomonas are good experimental organisms that can be manipulated at both the genetic and molecular levels, this means it should be feasible to discover the genetic innovations that made multicellularity possible within these species.
Multicellularity in the Volvocine Algae
How does Volvox compare to plants, animals, and other multicellular organisms with respect to the sorts of processes it has evolved? In a way, Volvox exhibits a relatively streamlined type of multicellularity. It possesses just two cell types, and these cells are not organized into tissues or organs. Nonetheless it has evolved an impressive degree of developmental and morphological novelty. Indeed, David Kirk compared the developmental programs of Volvox, Chlamydomonas, and several other volvocine algae and inferred that twelve new developmental traits evolved in Volvox that its unicellular ancestor did not possess (Kirk 2005). For instance, Volvox exhibits embryo inversion, a morphogenetic process that is analogous to animal gastrulation and that positions the flagellar ends of somatic cells correctly following cell division (Kirk & Nishii 2001). In addition, Volvox embryos execute a specialized type of cell division that generates cells of different sizes and types, called asymmetric division (Kirk 2001). And, as described above, Volvox makes specialized cell types: terminally differentiated somatic cells and immortal, stem cell-like gonidia. As we will see shortly, researchers have already discovered some clues about how each of these traits evolved.
Strategies for Investigating the Evolution of Multicellularity
How does one go about learning how multicellularity evolves? Most approaches that researchers have used to study the genetic basis of multicellularity fall into one of two strategies — comparative genomics and mutational/functional genetics — or a combination of the two. A third approach, involving molecular taxonomy studies that tell us about the relatedness of species, has also been very important. In fact, this third approach forms the foundation on which the other two approaches are built. However, it will not be discussed here, because it is beyond the scope of the current discussion.
With comparative genomic approaches, researchers compare fully sequenced genomes of close multicellular-unicellular cousins to determine which genes are unique to either genome, and to determine how the proteins encoded by the genomes differ. The idea with this type of analysis is that any genes or gene families present in the multicellular species but not the unicellular one might have been important for the evolution of multicellularity. That is, certain "special" genes for multicellularity might be found only in multicellular organisms. Or if the multicellular species contains significantly more copies of a certain kind of gene than does its unicellular cousin, or if the proteins encoded by certain related genes have changed a great deal in the two species, then those extra copies or changed proteins might be important for multicellularity.
It is important to keep in mind here that large-scale comparative genomic studies typically uncover only big differences in gene families, or differences in well-known genes and gene families. Such studies might not uncover subtle differences, such as small changes in the sizes of gene families that occur when a gene is duplicated (or lost) in one species but not the other. Evolutionary biologists think that gene duplication events could be extremely important for the evolution of new traits, because the new genes are free to change over time and subsequently function somewhat differently from the genes they were copied from.
Researchers also use mutational/functional approaches, which start by identifying mutant versions of the multicellular species that are defective for key developmental processes that don't occur in the unicellular species (such as the ability to make different cell types). These mutants are then used to clone the affected genes. After that, researchers analyze the unicellular species genome to determine whether the same (orthologous) genes exists and, if so, whether or how they differ from the multicellular versions. These types of investigations using current, living organisms are very powerful. They reveal valuable clues to which genes were important for evolving novel abilities, and how those genes were shuffled around and/or changed during the evolutionary past. On the whole, sorting out the differences between multicellular and unicellular organisms lends clues to how multicellularity may have evolved.
Lessons Learned from the Volvocine Algae: Comparative Genomics
What have the volvocine algae taught us about how multicellularity evolves? Recently researchers sequenced and compared the Chlamydomonas and Volvox genomes and found them to be remarkably similar (Prochnik et al. 2010). By almost every measure — overall genome size, number of protein-coding genes, number of different kinds of protein domains encoded, and distribution of gene family sizes — the two organisms are very much the same. When these investigators looked carefully at certain families of genes, especially those known to be involved in regulating the sorts of developmental processes that occur in Volvox but not Chlamydomonas, they again found only similarities, for the most part. They did find one very obvious and important difference, however: Compared to Chlamydomonas, Volvox has many more genes that encode cell wall/ECM proteins, and many of the extra genes are quite different from the ones Chlamydomonas has. Here it is important to point out that the cell wall surrounding Chlamydomonas has two parts: an inner layer and an outer one. Volvox has versions of both, but the inner layer is greatly expanded compared to the Chlamydomonas inner layer. It makes up the bulk of the ECM that is not present in Chlamydomonas, and it helps cement the Volvox cells together. Researchers believe that the explosion in cell wall genes, and the morphing of some of those genes into different kinds of cell wall genes, is what drove the creation of ECM in Volvox.
Lessons Learned from the Volvocine Algae: Mutational/Functional Genetics
Clearly, pure comparative genomic approaches have their limitations; they cannot tell us everything there is to know about how developmental processes and multicellularity evolve. But genetic screens are possible for Volvox and Chlamydomonas. What insights have these screens provided into how multicellularity evolved in the volvocine lineage?
Researchers have used genetic screens for developmental defects in Volvox to identify one gene that is essential for asymmetric division (glsA) and three others that are required for embryo inversion (invA, invB, and invC; Miller & Kirk 1999; Nishii et al. 2003; Ueki & Nishii 2008, 2009). All four genes have easily recognizable orthologs in Chlamydomonas that are very similar to their Volvox counterparts. Researchers have cloned Chlamydomonas orthologs corresponding to two of the Volvox developmental genes. One set of investigators showed that the GAR1 gene of Chlamydomonas, which is orthologous to glsA, is able to function just like glsA: When transformed into glsA mutants, it repaired, or rescued, their asymmetric division defect (Cheng et al. 2003). Likewise, another set of researchers found that IAR1 (orthologous to invA) can rescue the inversion defect of invA mutants (Nishii et al. 2003). These results tell us that the glsA/GAR1 and invA/IAR1 genes have not changed in important ways since the time that Volvox and Chlamydomonas diverged from a common ancestor.
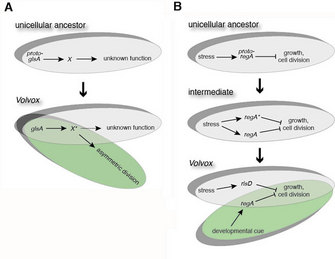
Additional insights of a different sort have come from analysis of the somatic regenerator, or regA, gene. This gene is required for maintenance of the somatic cell fate in Volvox; regA mutant somatic cells develop normally at first, but instead of remaining somatic cells their entire lives and then eventually dying, as somatic cells usually do, they enlarge and regenerate as gonidia that eventually divide to produce new spheroids (Kirk 1998). Therefore regA somehow prevents somatic cells from growing and dividing, and keeps them from having the stem cell-like potential that gonidia possess. Think of regA as a tumor suppressor gene that prevents the sort of uncontrolled growth that cancer cells exhibit. On analyzing the Volvox and Chlamydomonas genomes to determine how many regA-like genes they have, investigators discovered that both algae have a large family of paralogous genes that encode proteins resembling the regA product. But using phylogenetic analyses and other methods, they also found that Chlamydomonas does not have a regA gene (Duncan et al. 2007). Why not? In addition, where did regA come from in the first place, and how did it come to take on its role as a master regulator of the somatic cell fate?
Researchers found answers to some of these questions through further archaeological analysis of the Chlamydomonas and Volvox genomes. Their analyses revealed that regA likely was generated when a progenitor gene in the ancestor of Chlamydomonas and Volvox was inadvertently copied to produce two paralogous genes: one that eventually gave rise to regA, and one that gave rise to a related gene. While Volvox retained both regA and the other gene (a paralog), Chlamydomonas lost regA. In terms of how the regA function evolved, the modern-day versions of that other gene offer the best place to look for clues. Investigators studying this question found that the Chlamydomonas version of that regA-like gene, named RLS1, is turned on when Chlamydomonas is deprived of light or certain nutrients (Nedelcu 2009). This correlation suggests that perhaps RLS1 functions when cells are deprived of energy or nutrients. Since regA represses reproduction, it seems logical that RLS1 probably does too. If this is the case, then the capacity to make different cell types may have evolved from a pathway that repressed growth and cell division in response to energy/nutrient deprivation. This could have happened when the gene that controls that pathway was copied and then used to co-opt the entire pathway to repress growth and division in a developmental context (Figure 3B). Think of the hybrid car analogy again, except in this case the entire stress response pathway is the brake system. Something like this — the co-option of an existing genetic pathway so that it causes a cell to do something it would ordinarily do only under different circumstances — might explain, in general, how organisms evolve new cell types.
Summary
What Volvox and Chlamydomonas have taught us so far is that multicellularity, at least certain aspects of it, can evolve through relatively minor modifications of the unicellular blueprint (see Figure 3). Presumably not just any unicellular blueprint will do; no doubt the unicellular ancestor of Volvox already had many of the requisite genetic and cell biological raw materials for multicellularity: a multiple fission cell division program, a cell wall that could be modified into ECM, and possibly a stress response pathway that could be adapted to repress growth and division of a subset of cells, causing them to lose the ability to reproduce. But there is still much to learn. What new gene functions evolved to permit the evolution of asymmetric division and inversion? How did the other novel developmental traits of Volvox evolve? And are there similarities between the way multicellularity evolved in the volvocine algae and the way it evolved in other kinds of organisms? With the rate of recent progress in this field, answers to these questions, and more, should be on their way soon.
References and Recommended Reading
Buss, L. The Evolution of Individuality. Princeton, NJ: Princeton University Press, 1987.
Cheng, Q. et al. The role of GlsA in the evolution of asymmetric cell division in the green alga Volvox carteri. Development Genes and Evolution 213, 328–335 (2003).
Duncan, L. et al. The VARL gene family and the evolutionary origins of the master cell-type regulatory gene, regA, in Volvox carteri. Journal of Molecular Evolution 65, 1–11 (2007).
Herron, M. D. et al. Triassic origin and early radiation of multicellular volvocine algae. PNAS 106, 3254–3258 (2009).
Herron, M. D. & Michod, R. E. Evolution of complexity in the volvocine algae: Transitions in individuality through Darwin's eye. Evolution 62, 436–451 (2008).
King, N. The unicellular ancestry of animal development. Developmental Cell 7, 313–325 (2004).
Kirk, D. L. Germ-soma differentiation in Volvox. Developmental Biology 238, 213–223 (2001).
Kirk, D. L. A twelve-step program for evolving multicellularity and a division of labor. BioEssays 27, 299–310 (2005).
Kirk, D. L. Volvox: The Molecular Genetic Origins of Multicellularity and Cellular Differentiation. Cambridge: Cambridge University Press, 1998.
Kirk, D. L. & Nishii, I. Volvox carteri as a model for studying the genetic and cytological control of morphogenesis. Development, Growth & Differentiation 43, 621–631 (2001).
Miller, S. M. & Kirk, D. L. glsA, a Volvox gene required for asymmetric division and germ cell specification, encodes a chaperone-like protein. Development 126, 649–658 (1999).
Nedelcu, A. M. Environmentally induced responses co-opted for reproductive altruism. Biology Letters 5, 805–808 (2009).
Nishii, I. et al. A kinesin, invA, plays an essential role in Volvox morphogenesis. Cell 113, 743–753 (2003).
Peterson, K. J. & Butterfield, N. J. Origin of the Eumetazoa: Testing ecological predictions of molecular clocks against the Proterozoic fossil record. PNAS 102, 9547–9552 (2005).
Prochnik, S. E. et al. Genomic analysis of organismal complexity in the multicellular green alga Volvox carteri. Science 329, 223–226 (2010.)
Sanderson, M. J. Molecular data from 27 proteins do not support a Precambrian origin of land plants. American Journal of Botany 90, 954–956 (2003).
Ueki, N. & Nishii, I. Controlled enlargement of the glycoprotein vesicle surrounding a Volvox embryo requires the InvB nucleotide-sugar transporter and is required for normal morphogenesis. Plant Cell 21, 1166–1181 (2009).
Ueki, N. & Nishii, I. Idaten is a new cold-inducible transposon of Volvox carteri that can be used for tagging developmentally important genes. Genetics 180, 1343–1353 (2008).



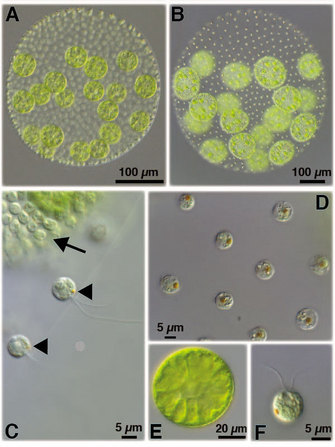
 Figure 2: Volvox carteri and Chlamydomonas reinhardtii
Figure 2: Volvox carteri and Chlamydomonas reinhardtii