« Prev Next »
Comparative genomics, which involves analysis of the nucleotide sequences of genomes, shows that the known life-forms comprise two major divisions: the cellular and the viral "empires." The cellular empire consists of three domains: Bacteria, Archaea, and Eukarya. What are the evolutionary relationships between the two empires and the three domains? Comparative genomics sheds light on this key question by showing that the previous conception of the Tree of Life should be replaced by a complex network of treelike and netlike routes of evolution to depict the history of life. Even under this new perspective on evolution, the two empires and the three cellular domains remain distinct. Furthermore, comparative genomics suggests that eukaryotes are archaebacterial chimeras, which evolved as a result of, or at least under a strong influence of, an endosymbiotic event that gave rise to mitochondria.
Cells, Viruses, and the Classification of Organisms
All living organisms consist of elementary units called cells. Cells are membrane-enclosed compartments that contain genomic DNA (chromosomes), molecular machinery for genome replication and expression, a translation system that makes proteins, metabolic and transport systems that supply monomers for these processes, and various regulatory systems. Scientists have performed careful microscopic observations and other experiments to show that all cells reproduce by different forms of division. Cell division is an elaborate process that ensures faithful segregation of copies of the replicated genome into daughter cells. The best-characterized cells are the relatively large cells of animals, plants, fungi, and diverse unicellular organisms known as protists, such as amoebae or paramecia. These cells possess an internal cytoskeleton and a complex system of intracellular membrane partitions, including the nucleus, a compartment that encloses the chromosomes. These organisms are known as eukaryotes because they possess a true nucleus (karyon in Greek). In contrast, the much smaller cells of bacteria have no nucleus and are named prokaryotes.
In the twentieth century, scientists devised new imaging methods like electron microscopy, which can be used to view tiny particles that are much smaller than cells, to detect a second fundamental form of biological organization: the viruses. Viruses are obligate intracellular parasites. These selfish genetic elements typically encode some proteins essential for viral replication, but they never contain the full complement of genes for the proteins and RNAs required for translation, membrane function, or metabolism. Therefore, viruses exploit cells to produce their components.
Classifying organisms (known as systematics or taxonomy) is one of the oldest occupations of biologists. Carolus Linnaeus constructed his now famous taxonomic system — certainly one of the foundations of scientific biology — in the middle of the eighteenth century. How did he classify organisms? Since Linnaeus was not an evolutionist, his classifications strived to reflect only similarities between species that were considered immutable. The goals of systematics changed after Charles Darwin introduced the concept of the Tree of Life (hereafter, TOL). At least in principle, the TOL was perceived as an accurate depiction of the evolutionary relationships between all life-forms. After Darwin, evolutionary biologists attempted to delineate monophyletic taxa, which are groups of organisms that share a common ancestry and thus form a distinct branch in the TOL. Until the last quarter of the twentieth century, however, taxonomists worked with phenotypic similarities between organisms, so monophyly remained a hypothesis based on the hierarchy of similar features. Accordingly, biologists could boast substantial advances in the classification of animals and plants, and to a lesser extent, simpler multicellular life-forms, such as fungi and algae. However, taxonomy was nearly helpless when it came to unicellular organisms, particularly bacteria, which have few easily observed features to compare. As a result, microbiologists were skeptical about whether it was possible to establish the evolutionary relationships between microbes. How could they compare these tiny organisms?
A revolution occurred in 1977 when Carl Woese and his co-workers performed pioneering studies to compare the nucleotide sequences of a molecule that is conserved in all cellular life-forms: the small subunit of ribosomal RNA (known as 16S rRNA). By comparing the nucleotide sequences of the 16S rRNA, they were able to derive a global phylogeny of cellular organisms for the first time. This phylogeny overturned the eukaryote-prokaryote dichotomy by showing that the 16S rRNA tree neatly divided into three major branches, which became known as the three domains of (cellular) life: Bacteria, Archaea and Eukarya (Woese et al. 1990). This discovery was enormously surprising, given that superficially the members of the new Archaea domain did not appear particularly different from bacteria. Since archaea and bacteria looked alike, how different could they be?
The Cellular Domains: Archaea, Bacteria, and Eukarya
With the progress of gene sequencing in the 1980s, many scientists performed phylogenetic studies to compare universally conserved proteins, such as protein subunits of the ribosome or of RNA polymerase. Their results supported the three-domain classification. Moreover, evolutionary biologists developed approaches to deduce the root position of the tree. Strikingly, they placed the LUCA between bacteria on one side and archaea together with eukaryotes on the other side, implying that archaea and eukaryotes share a common ancestor to the exclusion of bacteria (Figure 1b; Gogarten et al. 1989; Brown & Doolittle 1997). This finding emphasizes that similarity of cellular organization and common ancestry are two very different things.
The discovery of Archaea as a distinct, new domain of cellular life stimulated extensive studies into the molecular biology of these microbes, many of which thrive in unusual, extremely hot or salty environments. From these studies, researchers learned that the three domains are indeed fundamentally different at several cell biological levels, and not just in universal genes like the 16S rRNA. How do the domains of life differ? Scientists identified two key distinctions related to the DNA replication system and the membrane. The replication system of archaea is largely unrelated to that of bacteria, but it is homologous to the replication machinery of eukaryotes. Conversely, the archaeal membrane and the proteins involved in its formation are unique, whereas bacteria and eukaryotes share homologous membranes. Thus, archaea and bacteria differ with respect to the origin of some of their central cellular systems, whereas eukaryotes seem to combine important features of both archaea and bacteria.
Networks of Genome Evolution Replace the Tree of Life
Evolutionary biologists used the sequences of multiple genomes of diverse life-forms to construct and compare thousands of phylogenetic trees for individual genes. Unexpectedly, when comparing these trees they learned that genes generally have distinct evolutionary histories, and the trees built for different genes show different branching orders (topologies). The diversity of gene tree topologies is particularly pronounced among prokaryotes. For example, when scientists build trees for the numerous genes encoding metabolic enzymes or membrane transport proteins, the separation of archaea and bacteria is almost never precisely reproduced; instead, the archaeal and bacterial branches are mixed. This crucial finding indicates that genome evolution in prokaryotes is not a treelike process but is best represented by a complex network that combines treelike fragments corresponding to coherent evolution of multiple genes with numerous horizontal connections (Figure 1c; Doolittle & Bapteste 2007; Koonin & Wolf 2008).
What do these horizontal connections represent? They represent horizontal gene transfer (HGT), the exchange of genes between different species. Indeed, scientists have described mechanisms of HGT, even between archaea and bacteria. Numerous theoretical and experimental studies indicate that HGT is the principal mechanism of evolutionary innovation in prokaryotes (Pal et al. 2005). One well-known, medically important example is the spread of antibiotic resistance among pathogenic bacteria.
The importance and ubiquity of HGT notwithstanding, comprehensive comparative analyses of phylogenetic trees have shown that the treelike structure roughly corresponding to the rRNA phylogeny represents a central trend in the evolution of prokaryotes. These trees apparently reflect the concerted evolution of a core set of highly conserved, essential genes, most of which encode proteins involved in information transmission (Puigbo et al. 2009).
Symbiosis of Two Prokaryotic Cells at the Origin of Eukaryotes
In eukaryotes, HGT appears to be much less common than in prokaryotes. Nevertheless, eukaryotic genes seem to differ in their origins. The majority are most closely related to bacterial homologs, whereas a minority appear to be of archaeal origin (Esser et al. 2004). What purposes do these genes serve in eukaryotes? The "archaeal" genes in eukaryotes primarily, albeit not exclusively, encode proteins involved in information processing (translation, transcription, and replication). The "bacterial" genes encode mostly operational proteins, such as metabolic enzymes and membrane transporters.
Thus, eukaryotes are archaebacterial genetic chimeras; that is, they have combinations of genes from two very different organisms. How could eukaryotes have genes from two different organisms? The remarkable, unique process that explains this phenomenon is endosymbiosis, the invasion of one (host) cell by another, followed by degradation of the invader (endosymbiont), which becomes an organelle, like the mitochondrion. All known eukaryotic cells contain mitochondria, or related organelles, which play central roles in energy conversion. These mitochondria retain common features with bacterial cells, including a small genome and a mitochondrial translation system, which reveal beyond a doubt that they originated from a specific bacterial group, the a-proteobacteria. Many bacterial genes were transferred from the genome of the endosymbiont to the eukaryotic nuclear genome during evolution of the mitochondria. How do scientists believe this transfer could have occurred?
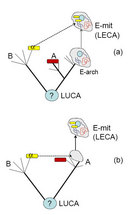
The connection between mitochondrial endosymbiosis and the origin of the signature features of the eukaryotic cell, such as the complex endomembrane system and cytoskeleton, remains a matter of debate (Embley & Martin 2006). One hypothesis holds that the host of the mitochondrial endosymbiont was a primitive eukaryotic cell (sometimes called an archaezoan) that possessed the signature structures of eukaryotes, including the nucleus, and was capable of phagocytosis (Figure 2a). The alternative hypothesis is that the host of the endosymbiont was an archaeon, and the endosymbiosis triggered the evolution of eukaryotic innovations (Figure 2b). Making a rigorous choice between the two hypotheses is extremely difficult. Unlike the archaezoan hypothesis, however, the endosymbiotic hypothesis accounts for the apparent lack of primitive amitochondrial eukaryotes that could be direct descendants of the archaezoa among the known eukaryotes. Furthermore, the endosymbiotic scenario proposes mechanistic causes for the origin of the intracellular structures, including the nucleus, in the emerging eukaryotic cell (Martin & Koonin 2006).
The World of Viruses
Scientists discovered viruses at the end of the nineteenth century as ultramicroscopic parasites of plants and animals, which passed through filters that held back bacteria. By the middle of the twentieth century, it became clear that viruses can replicate only within cells. However, the actual prominence of viruses in the biosphere and their role in the evolution of life were not revealed until the advances of metagenomics allowed for the massive sequencing of genes and genomes in environmental samples without the isolation of individual organisms. Viruses turn out to be the dominant biological entities on Earth. In the ocean, for example, viral particles outnumber cells by an order of magnitude (Suttle 2005). Viruses are also dominant in terms of genetic variety. Indeed, the greatest number of unique genes without detectable homologs in other genomes is found in viral genomes (Kristensen et al. 2010). In contrast with cellular life-forms — which all employ the same, classic strategy of DNA replication, transcription, and translation — viruses possess diverse genetic cycles. Viruses employ nearly all imaginable strategies of genome replication and expression: Some viruses have single-stranded or double-stranded RNA genomes that do not involve DNA in their replication, some have RNA genomes that use DNA as a replication intermediate, and some have genomes that are either single-stranded or double-stranded DNA molecules.
How do viral genomes compare to those of cellular life-forms? In comparison to cellular life-forms, viruses possess small genomes, ranging in size from between about 1,000 and 1,000,000 nucleotides. The genomes of giant viruses, such as the recently discovered Mimivirus that infects amoebae, are larger than the genomes of many bacteria and some archaea (Raoult & Forterre 2008). Viruses typically lack many of the genes that are universal among the three domains of cellular life — in particular, genes for translation system components. However, a small core of viral "hallmark genes" have been discovered that are missing in cellular life-forms. These genes encode proteins essential for virus reproduction (e.g., polymerases, helicases, and core virus particle components). These hallmark genes are shared by an extremely diverse group of viruses with different replication strategies, although none of the genes is strictly universal among viruses. The discovery of the hallmark genes reveals the evolutionary unity of the viral empire (Koonin et al. 2006).
Finally, viruses and related mobile genetic elements that lack capsids (e.g., plasmids, transposons, and others) are crucial for the evolution of cellular life-forms. These selfish genetic elements are major agents of gene transfer. The genomes of many eukaryotes, particularly animals and plants, consist in large part of inactivated remnants of such elements (up to 80 percent of the genome in plants).
Biologists sometimes debate whether viruses should be considered living organisms. The discovery of giant viruses like the Mimivirus have blurred the division between viruses and cells in terms of particle and genome size, leading to the revival of these debates (Moreira & López-Garcia 2009). However, the debates seem to be largely issues of semantics. Clearly, viruses constitute a distinct, major biological "empire" that is distinct from the empire of cellular life-forms, and the viral empire seems to eclipse the latter in terms of genetic complexity (Raoult & Forterre 2008).
Summary
Comparative genomics and metagenomics have transformed our understanding of the genetic universe. New discoveries have revealed the previously unrealized prominence of the viral world. This second biological empire seems to be even more vast and diverse than the empire of cellular life-forms. A second key transformation in our understanding is that a complex network of treelike and netlike routes better explains evolution than does a single TOL. Even under this new network perspective, the three domains of cellular life — Bacteria, Archaea, and Eukarya — remain objectively distinct. Although these domains are distinct, the eukaryotes are archaebacterial chimeras, which evolved as a result of, or at least under the strong influence of, an endosymbiotic event that gave rise to the mitochondria. Despite all the recent advances of evolutionary genomics, we still have to answer the most fundamental questions: How did cells evolve in the first place, what caused the fundamental differences between the two prokaryotic domains (Archaea and Bacteria), and what triggered the emergence of the complex organization of the eukaryotic cell?
References and Recommended Reading
Brown, J. R. & Doolittle, W. F. Archaea and the prokaryote-to-eukaryote transition. Microbiology and Molecular Biology Reviews 61, 456–502 (1997)
Doolittle, W. F. & Bapteste, E. Pattern pluralism and the Tree of Life hypothesis. PNAS 104, 2043–2049 (2007)
Embley, T. M. & Martin, W. Eukaryotic evolution, changes and challenges. Nature 440, 623–630 (2006)
Esser, C. et al. A genome phylogeny for mitochondria among alpha-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Molecular Biology and Evolution 21, 1643–1660 (2004)
Gogarten, J. P. et al. Evolution of the vacuolar H+-ATPase: Implications for the origin of eukaryotes. PNAS 86, 6661–6665 (1989)
Koonin, E. V., Senkevich, T. G. & Dolja, V. V. The ancient Virus World and evolution of cells. Biology Direct 19, 29 (2006)
Koonin, E. V. & Wolf, Y. I. Genomics of bacteria and archaea: The emerging dynamic view of the prokaryotic world. Nucleic Acids Research 36, 6688–6719 (2008)
Kristensen, D. M., et al. New dimensions of the virus world discovered through metagenomics. Trends in Microbiology 18, 11–19 (2010)
Martin, W., & Koonin, E. V. Introns and the origin of nucleus-cytosol compartmentation. Nature 440, 41–45 (2006)
Moreira, D., & López-García, P. Ten reasons to exclude viruses from the tree of
life. Nature Reviews Microbiology 7, 306–311 (2009)
Pal, C., Papp, B. & Lercher, M. J. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nature Genetics 37, 1372–1375 (2005)
Puigbo, P., Wolf, Y. I. & Koonin, E. V. Search for a Tree of Life in the thicket of the phylogenetic forest. Journal of Biology 8, 59 (2009)
Raoult, D. & Forterre, P. Redefining viruses: Lessons from mimivirus. Nature Reviews Microbiology 6, 315–319 (2008) doi:10.1038/nrmicro1858
Suttle, C. A. Viruses in the sea. Nature 437, 356–361 (2005)
Woese, C. R., Kandler, O. & Wheelis, M. L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. PNAS 87, 4576–4579 (1990)



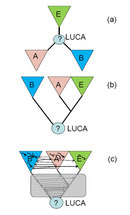
 Figure 1
Figure 1