« Prev Next »
Amino acids play a central role in cellular metabolism, and organisms need to synthesize most of them (Figure 1). Many of us become familiar with amino acids when we first learn about translation, the synthesis of protein from the nucleic acid code in mRNA. To date, scientists have discovered more than five hundred amino acids in nature, but only twenty-two participate in translation. In 1943, Gordon, Martin, and Synge used partition chromatography to separate and study constituents of proteins (Gordon, Martin, & Synge 1943), a major breakthrough that contributed to the rapid identification of the twenty amino acids used in proteins by all living organisms. After this initial burst of discovery, two additional amino acids, which are not used by all organisms, were added to the list: selenocysteine (Bock 2000) and pyrrolysine (Srinivasan et al. 2002).
Aside from their role in composing proteins, amino acids have many biologically important functions. They are also energy metabolites, and many of them are essential nutrients. Amino acids can often function as chemical messengers in communication between cells. For example, Arvid Carlsson discovered in 1957 that the amine 3-hydroxytyramine (dopamine) was not only a precursor for the synthesis of adrenaline from tyrosine, but is also a key neurotransmitter. Certain amino acids — such as citrulline and ornithine, which are intermediates in urea biosynthesis — are important intermediaries in various pathways involving nitrogenous metabolism. Although other amino acids are important in several pathways, S-adenosylmethionine acts as a universal methylating agent. What follows is a discussion of amino acids, their biosynthesis, and the evolution of their synthesis pathways, with a focus on tryptophan and lysine.
The Origins of Nutrient Biosynthesis
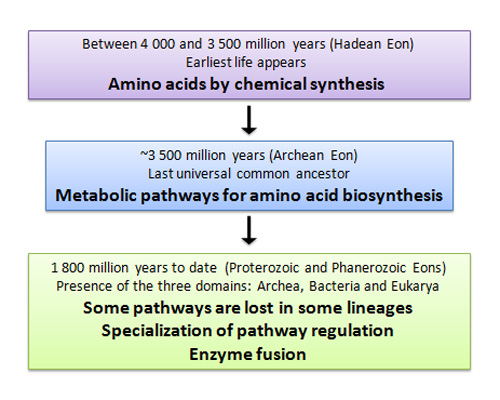
What Is an Amino Acid Made Of?
As implied by the root of the word (amine), the key atom in amino acid composition is nitrogen. The ultimate source of nitrogen for the biosynthesis of amino acids is atmospheric nitrogen (N2), a nearly inert gas. However, to be metabolically useful, atmospheric nitrogen must be reduced. This process, known as nitrogen fixation, occurs only in certain types of bacteria. Even though nitrogen is one of the most prominent chemical elements in living systems, N2 is almost unreactive (and very stable) because of its triple bond (N≡N). This bond is extremely difficult to break because the three chemical bonds need to be separated and bonded to different compounds. Nitrogenase is the only family of enzymes capable of breaking this bond (i.e., it carries out nitrogen fixation). These proteins use a collection of metal ions as the electron carriers that are responsible for the reduction of N2 to NH3. All organisms can then use this reduced nitrogen (NH3) to make amino acids. In humans, reduced nitrogen enters the physiological system in dietary sources containing amino acids. All organisms contain the enzymes glutamate dehydrogenase and glutamine synthetase, which convert ammonia to glutamate and glutamine, respectively. Amino and amide groups from these two compounds can then be transferred to other carbon backbones by transamination and transamidation reactions to make amino acids. Interestingly, glutamine is the universal donor of amine groups for the formation of many other amino acids as well as many biosynthetic products. Glutamine is also a key metabolite for ammonia storage. All amino acids, with the exception of proline, have a primary amino group (NH2) and a carboxylic acid (COOH) group. They are distinguished from one another primarily by , appendages to the central carbon atom.
Amino Acid Precursors and Biosynthesis Pathways
What Makes an Amino Acid Essential?
Not all the organisms are capable of synthesizing all the amino acids, and many are synthesized by pathways that are present only in certain plants and bacteria. Mammals, for example, must obtain eight of twenty amino acids from their diets. This requirement leads to a convention that divides amino acids into two categories: essential and nonessential (given a certain metabolism). Because of particular structural features, essential amino acids cannot be synthesized by mammalian enzymes (Reeds 2000). Nonessential amino acids, therefore, can be synthesized by nearly all organisms. The loss of the ability to synthesize essential amino acids likely emerged very early in evolution, because this dependence on other organisms for the source of amino acids is common among all eukaryotes, not just those of mammals.
How do certain amino acids become essential for a given organism? Studies in ecology and evolution give some clues. Organisms evolve under environmental constraints, which are dynamic over time. If an amino acid is available for uptake, the selective pressure to keep intact the genes responsible for that pathway might be lowered, because they would not be constantly expressing these biosynthetic genes. Without the selective pressure, the biosynthetic routes might be lost or the gene could allow mutations that would lead to a diversification of the enzyme's function. Following this logic, amino acids that are essential for certain organisms might not be essential for other organisms subjected to different selection pressures. For example, in 2000, Ishikawa and colleagues completed the genome sequence of the endosymbiont bacteria Buchnera, and in it they found the genes for the biosynthetic pathways necessary for the synthesizing essential amino acids for its symbiotic host, the aphid. Interestingly, those genes for the synthesis of its "nonessential" amino acids are almost completely missing (Shigenobu et al. 2000). In this way, Buchnera provides the host with some amino acids and obtains the other amino acids from the host (Baumann 2005; Pal et al. 2006).
Tryptophan Synthesis: Only Created Once
Free-living bacteria synthesize tryptophan (Trp), which is an essential amino acid for mammals, some plants, and lower eukaryotes. The Trp synthesis pathway appears to be highly conserved, and the enzymes needed to synthesize tryptophan are widely distributed across the three domains of life. This pathway is one of three that compose aromatic amino acids from chorismate (Figure 2, red pathway). (The other amino acids are phenylalanine and tyrosine.) Trp biosynthetic enzymes are widely distributed across the three domains of life (Xie et al. 2003). The genes that code for the enzymes in this pathway likely evolved once, and they did so more recently than those for other amino acid synthesis pathways. Researchers made this contention because all organisms containing this Trp synthesis pathway use homologous enzymes (Merino, Jensen, & Yanofsky 2008). As another point of distinction, the Trp pathway is the most biochemically expensive of the amino acid pathways, and for this reason it is expected to be tightly regulated.
Lysine Synthesis: Created Multiple Times
To date, scientists have discovered six different biosynthetic pathways in different organisms that synthesize lysine. These pathways can be grouped into the diaminopimelic acid (DAP) and aminoadipic acid (AAA) pathways (Figure 2, dark blue). The DAP pathway synthesizes lysine (Lys) from aspartate and pyruvate. Most bacteria, some archaea, fungi, algae, and plants use the DAP pathways. On the other hand, the AAA pathways synthesize Lys from alpha-ketoglutarate and acetyl coenzyme A. Most fungi, some algae, and some archaea use this route. Why do we observe this diversity, and why does it occur particularly for Lys synthesis? Interestingly, the DAP pathways retain duplicated genes from the biosynthesis of arginine, whereas the AAA pathways retain duplicated genes from leucine biosynthesis (Figure 2), indicating that each of the pathways experienced at least one duplication event during evolution (Hernandez-Montes et al. 2008; Velasco et al. 2002). Fani and coworkers performed a comparative analysis of the synthesis enzyme sequences and their phylogenetic distribution that suggested that the synthesis of leucine, lysine, and arginine were initially carried out with the same set of versatile enzymes. Over the course of time came a series of gene duplication events and enzyme specializations that gave rise to the unambiguous pathways we know today. Which of the pathways appeared earlier is still a source of query and debate.
To support this hypothesis, there is evidence from a fascinating archaea, Pyrococcus horikoshii. This organism can synthesize leucine, lysine, and arginine, yet its genome contains only genes for one pathway. Such a gap indicates that P. horikoshii has a mechanism similar to the ancestral one: versatile enzymes. Biochemical experiments are needed to further support the idea that these enzymes can use multiple substrates and to rule out the possibility that amino acid synthesis in this organism does not arise from enzymes yet unidentified.
Synthesis on the tRNA molecule
Selenocysteine (SeC) (Bock 2000) is a genetically encoded amino acid not present in all organisms. Scientists have identified SeC in several archaeal, bacterial, and eukaryotic species (even mammals). When present, SeC is usually confined to active sites of proteins involved in reduction-oxidation (redox) reactions. It is highly reactive and has catalytic advantages over cysteine, but this high reactivity is undermined by its potential to cause cell damage if free in the cytoplasm. Hence, it is too dangerous, and no pool of free SeC is available. How, then, is this amino acid synthesized for use in protein synthesis? The answer demonstrates the versatility of synthesis strategies deployed by organisms forced to cope with singularities. The synthesis of SeC is carried out directly on the tRNA substrate before being used in protein synthesis. First, SeC-specific tRNA (tRNAsec) is charged with serine via seril-tRNA synthetase, which acts in a somehow promiscuous fashion, serilating either tRNAser or tRNAsec. Then, another enzyme modifies Ser to SeC by substituting the OH radical with SeH, using selenophosphate as the selenium donor (Figure 2, pink pathway). This synthesis is a form of a trick to avoid the existence of a free pool of SeC while still maintaining a source of SeC-tRNAsec needed for protein synthesis. Strictly speaking, this mechanism is not an actual synthesis of amino acids, but rather a synthesis of aminoacetylated-tRNAs. However, this technique involving tRNA directly is not exclusive to SeC, and similar mechanisms dependent on tRNA have been described for asparagine, glutamine, and cysteine. Owing to its appearance of SeC across all three domains of life, scientists wonder if it is an ancestral mechanism for amino acid biosynthesis or simply a coincidence of selection pressures.
How Do Metabolic Pathways Evolve? Two Different Models
In 1945, Horowitz proposed the first accepted model for metabolic pathway evolution (Horowitz 1945). Called the retrograde model, it states that after an enzyme consumes all its substrate available, another enzyme capable of producing the aforementioned substrate is required, so the last enzyme evolved to the preceding one by a gene duplication and selection mechanism. In other words, enzymes evolve from others with similar substrate specificity, and the substrate of the last enzyme is the product of the preceding one. Also, the active site must bind both the substrate and the product. This model became very popular, but as more genes have been sequenced and more phylogenetic analyses performed, this mechanism has become less seemingly plausible and therefore unpopular. An alternative model, the patchwork assembly model, proposes that ancestral enzymes were generalists, so they could bind a number of substrates to carry out the same type of reaction. Gene duplication events followed by evolutionary divergence would result in enzymes with high affinity and specificity for a substrate. In other words, enzymes are recruited from others with the same type of chemical reaction. Whole genome analysis of Escherichia coli supports the patchwork evolution model (Teichmann et al. 2001). Duplication of whole pathways does not occur very often; nevertheless, examples include tryptophan (to synthesize paraminobenzoate) and histidine (to synthesize nucleotides) biosynthesis, as well as lysine, arginine, and leucine biosynthesis (see aforementioned example).
Other mechanisms, such as gene fusion, might occur in the process of pathway evolution. When gene fusions occur between the genes for different proteins of the same pathway, a mechanism that facilitates ligand binding is provided because the substrate of one domain is the product of the other; thus, passive diffusion becomes unnecessary. Fusions can also result in the tight regulation of fused domains. Histidine biosynthesis is a good example of gene fusion; at least seven genes of this pathway underwent fusion events in different phylogenetic lineages. This assertion means that fusions must be relatively recent because they occurred after the lineages arose (Fani et al. 2007). Another important pathway evolution mechanism is horizontal gene transfer, which allows the rapid acquisition of fully functional enzymes and pathways.
Open Questions about Amino Acid Evolution
Amino acids are one of the first organic molecules to appear on Earth. As the building blocks of proteins, amino acids are linked to almost every life process, but they also have key roles as precursor compounds in many physiological processes. These processes include intermediary metabolism (connections between carbohydrates and lipids), signal transduction, and neurotransmission. Recent years have seen great advances in understanding amino acid evolution, yet many questions on the subject of amino acid synthesis remain. What was the order of appearance of amino acids over evolutionary history? How many amino acids are used in protein synthesis today? How many were present when life began? Were there initially more than twenty used for building blocks, but intense selective process streamlined them down to twenty? Conversely, was the initial set much less than twenty, and did new amino acids successively emerge over time to fit into the protein synthesis repertoire? What are the tempo and mode of amino acid pathway evolution? These questions are waiting to be tackled — with old or new hypotheses, conceptual tools, and methodological tools — and are ripe for a new generation of scientists.
Summary
Scientists now recognize twenty-two amino acids as the building blocks of proteins: the twenty common ones and two more, selenocysteine and pyrrolysine. Amino acids have several functions. Their primary function is to act as the monomer unit in protein synthesis. They can also be used as substrates for biosynthetic reactions; the nucleotide bases and a number of hormones and neurotransmitters are derived from amino acids. Amino acids can be synthesized from glycolytic or Krebs cycle intermediates. The essential amino acids, those that are needed in the diet, require more steps to be synthesized. Some amino acids need to be synthesized when charged onto their corresponding tRNAs. We have discussed only two biosynthetic routes: the Trp pathway, which appears to have evolved only once, and the Lys pathway, which seems to have evolved independently in different lineages. Prevailing evidence suggests that metabolic pathways themselves seem to be evolving following the patchwork assembly model, which proposes that pathways originated through the recruitment of generalist enzymes that could react with a wide range of substrates. The study of the evolution of amino acid metabolism has helped us understand the evolution of metabolism in general.
References and Recommended Reading
Baumann, P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annual Review of Microbiology 59, 155–189 (2005) doi:10.1146/annurev.micro.59.030804.121041.
Bock, A. Biosynthesis of selenoproteins — an overview. Biofactors 11, 77–78 (2000).
Fani, R. et al. The role of gene fusions in the evolution of metabolic pathways: The histidine biosynthesis case. BMC Evolutionary Biology 7 Suppl 2, S4 (2007) doi:10.1186/1471-2148-7-S2-S4.
Gordon, A. H., Martin, A. J. & Synge, R. L. Partition chromatography in the study of protein constituents. Biochemical Journal 37, 79–86 (1943).
Hernandez-Montes, G. et al. The hidden universal distribution of amino acid biosynthetic networks: A genomic perspective on their origins and evolution. Genome Biology 9, R95 (2008) doi:10.1186/gb-2008-9-6-r95.
Horowitz, N. H. On the evolution of biochemical syntheses. Proceedings of the National Academy of Sciences 31, 153-157 (1945).
Merino, E., Jensen, R. A. & Yanofsky, C. Evolution of bacterial trp operons and their regulation. Current Opinion in Microbiology 11, 78–86 (2008) doi:10.1016/j.mib.2008.02.005.
Miller, S. L. A production of amino acids under possible primitive earth conditions. Science 117, 528–529 (1953).
Pal, C. et al. Chance and necessity in the evolution of minimal metabolic networks. Nature 440, 667–670 (2006) doi:10.1038/nature04568.
Reeds, P. J. Dispensable and indispensable amino acids for humans. Journal of Nutrition 130, 1835S–1840S (2000).
Shigenobu, S. et al. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature 407, 81–86 (2000) doi:10.1038/ng986.
Srinivasan, G., James, C. M. & Krzycki, J. A. Pyrrolysine encoded by UAG in archaea: Charging of a UAG-decoding specialized tRNA. Science 296, 1459–1462 (2002) doi:10.1126/science.1069588.
Teichmann, S. A. et al. The evolution and structural anatomy of the small molecule metabolic pathways in Escherichia coli. Journal of Molecular Biology 311, 693–708 (2001) doi:10.1006/jmbi.2001.4912.
Velasco, A. M., Leguina, J. I. & Lazcano, A. Molecular evolution of the lysine biosynthetic pathways. Journal of Molecular Evolution 55, 445–459 (2002) doi:10.1007/s00239-002-2340-2.
Xie, G. et al. Ancient origin of the tryptophan operon and the dynamics of evolutionary change. Microbiology and Molecular Biology Reviews 67, 303–342 (2003) doi:10.1128/MMBR.67.3.303-342.2003.



 Figure 2
Figure 2


























