« Prev Next »
New gene origination is a driving force of evolutionary innovation in all organisms. Recent research has focused on identifying the mechanisms that generate new genes, and scientists have found that these mechanisms involve a variety of molecular events, all of which must occur in the germ line to be inherited by the next generation. After the germ-line mutational event, the new gene (e.g., a new gene duplicate located on human chromosome 2) will be polymorphic in the population; in other words, not all second chromosomes in the population will carry the duplication. Subsequently, the two most likely outcomes for the new gene are fixation (i.e., the new gene will reach a frequency of 100%) or extinction (i.e., the new gene will be lost).
Current knowledge of the origin of new genes encompasses information regarding both protein-coding genes and RNA genes. All of these genes are transcribed, but only protein-coding genes are translated into proteins. The study of pseudogenes, originally defined as sequences that resemble known genes but cannot produce a functional protein, has revealed not only how often genes degenerate, but also that many sequences once believed to be degenerating protein-coding genes are in fact functional RNA genes.
Mechanisms of New Gene Generation
Over the years, scientists have proposed several mechanisms by which new genes are generated. These include gene duplication, transposable element protein domestication, lateral gene transfer, gene fusion, gene fission, and de novo origination.
Gene Duplication
Gene duplication was the first mechanism of gene generation to be suggested (Ohno, 1970), and this process does indeed appear to be the most common way of creating new genes. Duplications are typically classified according to the size of the portion of the genome that is duplicated; thus, a duplication may be described as involving an entire genome, large segments of a genome, individual genes, individual exons, or even specific parts of exons (Betrán & Long, 2002). The mechanisms that generate duplicate genes are diverse, and more details about these mechanisms are continually being discovered. These mechanisms include whole-genome duplications originating through nondisjunction, tandem duplications originating through unequal crossover, retropositions originating through the retrotranscription of an RNA intermediate, transpositions involving transposable elements (Jiang et al., 2004; Morgante et al., 2005), and duplications occurring after rearrangements and subsequent repair of staggered breaks (Ranz et al., 2007). Such duplications involve not only protein-coding genes, but also noncoding RNA genes. For example, a novel class of retroduplicates includes snoRNAs, which are a class of RNA genes that are involved in ribosomal RNA processing (Weber, 2006).
Much of the current excitement about gene duplication stems from the fact that with the number of sequenced genomes now available, researchers have more accurate estimates of how often genes duplicate, and these rates are extremely high. For example, more than 100 genes duplicate in the human genome per 1 million years (Hahn et al., 2007a). This means that the percentage of the genome affected by gene number differences (estimated to be 6%) contributes more to the differences between humans and chimpanzees than do single nucleotide differences between orthologous sequences (estimated to be 1.5% [Demuth et al., 2006]). High rates (17 genes per 1 million years) have also been estimated in flies (Hahn et al., 2007b). Additional excitement comes from the realization that duplications occur so often that individuals of the same species differ greatly in DNA content and gene number (i.e., many duplications are polymorphic and contribute to individual differences [Sebat et al., 2004]). It is estimated that, on average, two humans will differ by approximately 5 megabases of information.
Unexpectedly, several duplication trends have been described in genomes with respect to sex chromosome evolution. Many new male genes originate in species' Y chromosomes. Some of these male genes are organized in families that undergo gene conversion to avoid Y-chromosome degeneration. Male germ-line genes can also duplicate out of the X chromosome through retroposition (Betrán et al., 2002; Emerson et al., 2004; Lahn et al., 2001; Rozen et al., 2003). These findings reveal that genomic location and organization matter for gene origination and function.
Transposable Element Protein Domestication
Transposable elements (TEs) are so-called "selfish" segments of DNA that encode proteins that allow these segments to copy or move themselves within a genome. There are two types of TEs: DNA transposons and retrotransposons. DNA transposons are able to excise themselves out of the genome and be inserted somewhere else, whereas retrotransposons copy themselves through an RNA intermediate. Similar to viral insertions in the genome, TE insertions cause mutations and contribute to increased genome size, but they usually do not encode cellular proteins.
Interestingly, one way for a genome to acquire new genes is by recruiting transposable element proteins and using them as cellular proteins. Such events are called domestications of TE proteins. A recent review by Fechotte and Pritham (2007) compiled many examples of these events, and it revealed that some of the unexpected functions in which TE domesticated proteins play a role include functioning of the vertebrate immune system and light sensing in plants. Several examples of domestication have also been described in Drosophila (Casola et al., 2007). In one case, codomestication of the two proteins encoded by the PIF/Harbinger transposable element was observed. In this fascinating example, the two genes that the original TE contained were domesticated simultaneously; one of these genes encoded a transposase that binds and cuts DNA, while the other encoded a protein that contains a Myb/SANT domain believed to function in transcription, chromatin remodeling, and protein-protein interactions. More data are needed to reveal whether both of these proteins were domesticated to function in the same biological process.
Lateral Gene Transfer
Scientists use the term "lateral gene transfer" to refer to the case in which a gene does not have a vertical origin (i.e., direct inheritance from parent to offspring) but instead comes from an unrelated genome. It is well known that this sort of transfer occurs between bacteria, and that it also has taken place between the genomes of the cellular organelles (mitochondria and chloroplasts) and the nuclear genomes (Roger, 1999). However, more recent transfer events between organelles and/or endosymbiont bacteria continue to occur (Bergthorsson et al., 2003; Hotopp et al., 2007). For example, large-scale sequencing efforts have revealed that much of the genome of the intracellular endosymbiont Wolbachia pipentis was integrated into Drosophila species (Hotopp et al., 2007). However, the mechanism for these transfers remains largely unknown, and the functional consequences of some of these transfers have yet to be explored.
Gene Fusion and Fission
Existing genes can also fuse (i.e., two or more genes can become part of the same transcript) or undergo fission (i.e., a single transcript can break into two or more separate transcripts), thereby forming new genes. Interestingly, it has been observed that chimeric fusion genes sometimes involve two copies of the same gene (e.g., the alcohol dehydrogenase gene in Drosophila), and when that happens, the resulting genes undergo parallel evolution (Jones & Begun, 2005), in which they shift away from the functions of their parental genes.
De Novo Gene Origination
New genes can additionally originate de novo from noncoding regions of DNA. Indeed, several novel genes derived from noncoding DNA have recently been described in Drosophila (Begun et al., 2007; Levine et al., 2006). For these recently originated Drosophila genes with likely protein-coding abilities, there are no homologues in any other species. Note, however, that the de novo genes described in various species thus far include both protein-coding and noncoding genes. These new genes sometimes originate in the X chromosome, and they often have male germ-line functions.
The action of all the mechanisms described in the previous sections leads to exon shuffling (i.e., the observation that many genes share exons) (Gilbert et al., 1997; Li et al., 2001). In addition, analyses of "young" genes (genes that originated only a few million years ago) allow investigators to document all of the events that gave rise to these genes, because time has not eroded the footprints of these events. Using this approach, it has been inferred that combinations of several mechanisms and several events are often responsible for the generation of new genes (Betrán & Long, 2002). Two good examples of this are the jingwei gene (Long & Langley, 1993) and the SETMAR gene (Cordaux et al., 2006; Figure 1).
What Happens to New Genes?
All these new sequences add to the complexity and diversity of genomes. As with any mutation, when new genes become fixed in a genome, they add to the differences between species and serve as the raw material for evolution (Ohno, 1970). This is easy to see in the case of gene duplication. Gene duplication results in two or more copies of a gene: one that can maintain its original function in the organism, and other(s) that can be "played with" to take on new functions. As a consequence, new duplicates are a main source of genome innovation and often evolve under positive selection, in which rapid changes in the protein encoded by the new gene occur to gain a new function (Presgraves, 2005). This process is referred to as neofunctionalization of the new gene.
Other possible outcomes after duplication include gene loss or pseudogenization; maintenance of both genes as a way to increase expression or to maintain multiple variants within individuals (essentially "fixing" heterozygosity); or the occurrence of subfunctionalization (i.e., the occurrence of mutually complementing neutral disabling mutations such that both genes need to be kept in the genome [Lynch & Force, 2000]). Subfunctionalization is an interesting phenomenon because it begins with a partition of function but can set the grounds for specialization (Torgerson & Singh, 2004). Some mixed outcomes (such as subfunctionalization followed by neofunctionalization and subneofunctionalization) are also possible (He & Zhang, 2005).
One unanticipated consequence of gene duplication and gene loss is that these events can become the basis for some incompatibilities between species. Duplications and losses have been shown to play a role in hybrid breakdown and in the reduced fitness of the descendants of matings between genetically differentiated populations. Thus, these processes might contribute to the process of speciation (Masly et al., 2006).
The Origin and Fate of Pseudogenes
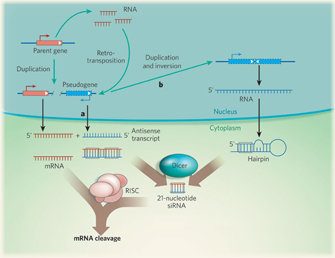
As previously mentioned, pseudogenes are commonly defined as sequences that resemble known genes but cannot produce functional proteins. Pseudogenes originate through the same mechanisms as protein-coding genes, followed by the subsequent accumulation of disabling mutations (e.g., nucleotide insertions, deletions, and/or substitutions) that disrupt the reading frame or lead to the insertion of a premature stop codon. Pseudogenes can be broadly classified into two categories: processed and nonprocessed. Nonprocessed pseudogenes usually contain introns, and they are often located next to their paralogous parent gene. Processed pseudogenes are thought to originate through retrotransposition; accordingly, they lack introns and a promoter region, but they often contain a polyadenylation signal and are flanked by direct repeats. Errors in reverse transcription and the lack of an appropriate regulatory environment often lead to the degeneration of processed copies of genes (D'Errico et al., 2004).
The abundance of pseudogenes in a given genome usually depends on rates of gene duplication and loss. Mammals appear to have a high number of processed pseudogenes—approximately 8,000 (Zhang et al., 2003; Zhang et al., 2004). On the other hand, most other organisms have many fewer; for instance, in Drosophila, only 20 retropseudogenes are detectable (Harrison et al., 2003). This pattern has been explained by the deletion bias that exists in Drosophila; indeed, after studying the size distribution of deletions in Drosophila and mammals, researchers concluded that deletions in Drosophila are much bigger (Petrov & Hartl, 2000).
The most interesting pseudogene finding to date is that degenerated protein-coding genes have been proven to "live on" as RNA genes (Sasidharan & Gerstein, 2008). Although researchers had previously proven that pseudogenes could be transcribed (Harrison et al., 2005), they only recently realized that these sequences can regulate parental genes through siRNAs; evidence of this phenomenon has been found in both flies and mammals (Figure 2; Sasidharan & Gerstein, 2008). In addition, some processed pseudogenes seem to have evolved into primate microRNA genes (Devor, 2006).
Of course, genomes remain full of surprises. Additional work will continue to reveal even more about the functional potential of many new DNA sequences.
References and Recommended Reading
Begun, D., et al. Evidence for de novo evolution of testis-expressed genes in the Drosophila yakuba/Drosophila erecta clade. Genetics 176, 1131–1137 (2007)
Bergthorsson, U., et al. Widespread horizontal transfer of mitochondrial genes in flowering plants. Nature 424, 197–201 (2003) (link to article)
Betrán, E., & Long, M. Expansion of genome coding regions by acquisition of new genes. Genetica 115, 65–80 (2002)
Betrán, E., et al. Retroposed new genes out of the X in Drosophila. Genome Research 12, 1854–1859 (2002)
Casola, C., et al. PIF-like transposons are common in Drosophila and have been repeatedly domesticated to generate new host genes. Molecular Biology and Evolution 24, 1872–1888 (2007)
Cordaux, R., et al. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proceedings of the National Academy of Sciences 103, 8101–8106 (2006)
Demuth, J., et al. The evolution of mammalian gene families. PLoS ONE 1, e85 (2006)
D'Errico, I., et al. Pseudogenes in metazoa: Origin and features. Briefings in Functional Genomics and Proteomics 3, 157–167 (2004)
Devor, E. J. Primate microRNAs miR-220 and miR-492 lie within processed pseudogenes. Journal of Heredity 97, 186–190 (2006)
Emerson, J. J., et al. Extensive gene traffic on the mammalian X chromosome. Science 303, 537–540 (2004)
Fechotte, C., & Pritham, E. DNA transposons and the evolution of eukaryotic genomes. Annual Review of Genetics 41, 331–368 (2007)
Gilbert, W., et al. Origin of genes. Proceedings of the National Academy of Sciences 94, 7698–7703 (1997)
Hahn, M., et al. Accelerated rate of gene gain and loss in primates. Genetics 177, 1941–1949 (2007a)
Hahn, M., et al. Gene family evolution across 12 Drosophila genomes. PLoS Genetics 3, e197 (2007b)
Harrison, P. M., et al. Identification of pseudogenes in the Drosophila melanogaster genome. Nucleic Acids Research 31, 1033–1037 (2003)
Harrison, P. M., et al. Transcribed processed pseudogenes in the human genome: An intermediate form of expressed retrosequence lacking protein-coding ability. Nucleic Acids Research 33, 2374–2383 (2005)
He, X., & Zhang, J. Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169, 1157–1164 (2005)
Hotopp, J., et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317, 1753–1756 (2007)
Jiang, N., et al. Pack-MULE transposable elements mediate gene evolution in plants. Nature 431, 569–573 (2004) (link to article)
Jones, C. D., & Begun, D. J. Parallel evolution of chimeric fusion genes. Proceedings of the National Academy of Sciences 102, 11373–11378 (2005)
Lahn, B. T., Pearson, N. M., & Jegalian, K. The human Y chromosome, in the light of evolution. Nature Reviews Genetics 2, 207–216 (2001) (link to article)
Levine, M. T., et al. Novel genes derived from noncoding DNA in Drosophila melanogaster are frequently X-linked and exhibit testis-biased expression. Proceedings of the National Academy of Sciences 103, 9935–9939 (2006)
Li, W. H., et al. Evolutionary analyses of the human genome. Nature 409, 847–849 (2001) (link to article)
Long, M., & Langley, C. H. Natural selection and the origin of jingwei, a chimeric processed functional gene in Drosophila. Science 260, 91–95 (1993)
Lynch, M., & Force, A. The probability of duplicate gene preservation by subfunctionalization. Genetics 154, 459–473 (2000)
Masly, J., et al. Gene transposition as a cause of hybrid sterility in Drosophila. Science 313, 1448–1450 (2006)
Morgante, M., et al. Gene duplication and exon shuffling by helitron-like transposons generate intraspecies diversity in maize. Nature Genetics 37, 997–1002 (2005) (link to article)
Ohno, S. Evolution by Gene Duplication (Springer-Verlag, Berlin, 1970)
Petrov, D., & Hartl, D. Pseudogene evolution and natural selection for a compact genome. Journal of Heredity 91, 221–227 (2000)
Presgraves, D. C. Evolutionary genomics: New genes for new jobs. Current Biology 15, R52–R53 (2005)
Ranz, J., et al. Principles of genome evolution in the Drosophila melanogaster species group. PLoS Biology 5, e152 (2007)
Roger, A. Reconstructing early events in eukaryotic evolution. American Naturalist 154, S146–S163 (1999)
Rozen, S., et al. Abundant gene conversion between arms of palindromes in human and ape Y chromosomes. Nature 423, 873–876 (2003) (link to article)
Sasidharan, R., & Gerstein, M. Genomics: Protein fossils live on as RNA. Nature 453, 729–731 (2008) (link to article)
Sebat, J., et al. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004)
Torgerson, D. G., & Singh, R. S. Rapid evolution through gene duplication and subfunctionalization of the testes-specific alpha-4 proteasome subunits in Drosophila. Genetics 168, 1421–1432 (2004)
Weber, M. Mammalian small nucleolar RNAs are mobile genetic elements. PLoS Genetics 2, e205 (2006)
Zhang, Z., Carriero, N., & Gerstein, M. Comparative analysis of processed pseudogenes in the mouse and human genomes. Trends in Genetics 20, 62–67 (2004)
Zhang, Z., et al. Millions of years of evolution preserved: A comprehensive catalog of the processed pseudogenes in the human genome. Genome Research 13, 2541–2558 (2003)




 Figure 1
Figure 1


























