« Prev Next »
The truth about the platypus—and what makes the animal's recent genomic sequencing particularly interesting—is that it belongs to a lineage that separated from ours approximately 166 million years ago, deep in the Mesozoic era, and since that time, it has independently lost different elements of our last common ancestor. By comparing bits of the platypus genome that were conserved with those that were lost, researchers can develop a clearer picture of what Jurassic mammals were like, and they can also determine what sorts of genetic traits contemporary mammals have gained and lost over the course of evolution.
Are Shared Characteristics a Sign of Relatedness?
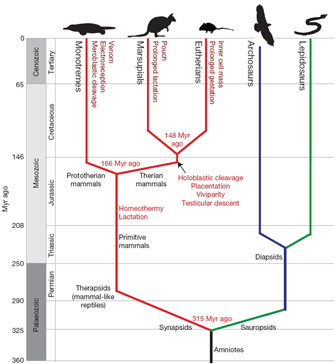
In order to better understand the impact of the platypus study, it is helpful to begin by looking at a cladogram that illustrates the dates when derived traits appeared in the various lineages considered in the study (Figure 1). This diagram reflects a fairly conventional picture of our evolutionary history, and it reinforces the evolutionary explanation for the illustrated relationships.
As you can see, a number of modern animals—including birds—are depicted along the top of the cladogram in Figure 1. Note, however, that the cladogram does not imply that modern monotremes (including the platypus) are part bird. Rather, birds are included in this diagram because they are contemporary representatives of the sauropsid lineage, a group of reptile cousins that split from our family tree roughly 315 million years ago. So, if monotremes aren't part bird, why did the investigators who conducted the platypus study examine genomic data from chickens during the course of their research? The primary reason for inclusion of this data relates to comparison. Specifically, the researchers knew that if they found a feature in birds that was also present in monotremes (or marsupials or eutherians), this would mean that the feature was most likely also present in the animals' common ancestor.
For instance, one of the unusual (for a mammal) features of the platypus is meroblastic cleavage. In fact, there is a famous telegram from 1884 in which researchers who were working in Australia announced a dramatic discovery to their colleagues at the British Association: "Monotremes oviparous, ovum meroblastic." Those four words declare that the platypus lays eggs (i.e., it is oviparous) and that the early stages of platypus embryo formation resemble the stages seen in birds and reptiles, not mammals. As opposed to the platypus, eutherians and marsupials have eggs that go through holoblastic cleavage; this means that the first cell divisions following fertilization cut all the way through the ovum, producing multiple, separable daughter cells. On the other hand, in the meroblastic cleavage of the platypus and chicken, the large yolky egg does not completely subdivide, so early cell divisions are incomplete. These incomplete divisions produce a sheet of cells on top of the yolk that are cytoplasmically continuous with the yolk cytoplasm. Indeed, this sheet is a common feature in yolky eggs and is a consequence of physical constraints on cell division.
Thus, both the platypus and the chicken exhibit meroblastic cleavage—but this does not mean that platypuses are part bird. Rather, what it does suggest is that meroblastic cleavage is likely a primitive character, one that was inherited from the last common ancestor of synapsids and sauropsids over 300 million years ago. Another possibility is that birds and monotremes evolved this feature independently, thereby making this trait an example of convergent evolution. Simple observation of meroblastic cleavage in both monotremes and birds is not enough to determine whether this characteristic arose via convergence or via common descent—in order to discover which is the case, we must look at multiple details of the evolutionary process.
Of course, not just the platypus but every living organism is a mix of both conserved, primitive characters and evolutionary novelties—thus, a mouse is just as "weird" as a platypus from an evolutionary perspective, as each is the product of processes that promoted divergence from a common ancestor, and each is equidistant from that ancestor. It's just that we primates share more derived characters, or synapomorphies, with mice than with platypuses because we are more closely related, and the mix of characters in mice is therefore more familiar to us.
Indeed, all modern animals are products of different evolutionary trajectories, and no one species by itself is representative of the ancestral condition. As a result, we must determine the ancestral state of modern animals through the comparison of multiple lineages—and that is the virtue of the data from the platypus. Information regarding the platypus genome adds yet another lineage to our data set—a lineage that diverged from ours over 160 million years ago. The platypus data is therefore a lens that can help us see what novelties arose during that 160 million year window on both the eutherian and monotreme sides of the split.
Lessons Learned from the Platypus
So, what are some of the details that we've learned from the platypus? One important message relates to the unity of life. Sequencing of the platypus genome reveals that the platypus has about 18,000 genes; humans, by comparison, have somewhere around 20,000. Moreover, roughly 82% of the platypus's genes are shared between monotremes, marsupials, eutherians, birds, and reptiles. This is not at all surprising, because all of these organisms are made of eukaryotic cells, and the basic eukaryotic machinery is going to be shared among species. Platypuses and humans also share a lot of "selfish" DNA bits—about half of both species' genomes consists of LINE and SINE-like sequences.
Humans and platypuses do differ in the details, however. For instance, an obvious difference is that the platypus lays yolky eggs, whereas humans and other eutherians have yolkless eggs that are retained in the mother's body. Thus, as you might expect, the platypus has a gene that humans lack—one that codes for vitellogenin, a crucial yolk protein.
As opposed to the presence of vitellogenin, a trait that both eutherians and monotremes have in common—but one that is not shared with birds—is lactation. (Although some birds can produce crop milk, this is a different adaptation). In the ancestral state, lactation was probably the secretion of fluids and immune system proteins to keep eggs and newborns hydrated and protected, but in our history, parents who invested more effort in secreting additional nutritive components, like sugars, fats, proteins, and calcium, were more successful. Like humans, the platypus secretes a true milk that is loaded with all of these components, including a protein called casein, which is thought to have originated by way of the duplication of a tooth enamel matrix protein gene, of all things. Today, two genes that code for proteins related to tooth production (enamelin and ameloblastin) are clustered with the casein-producing gene in both the platypus and the mouse, suggesting that the kind of sophisticated lactation abilities shared by monotremes and eutherians arose prior to the Jurassic period.
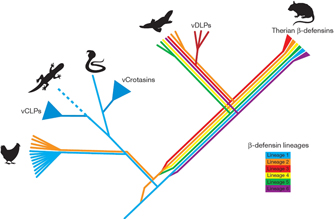
One particularly interesting specialization in the platypus is the evolution of venoms. The platypus has small, sharp spurs on its hind limbs that it uses to inject defensive poisons into predators, an unusual feature not found in other mammals. Where did these venoms come from? As it turns out, they arose through the duplication of genes that have other functions, with subsequent divergence. Many of these genes are involved in the functioning of the platypus's innate immune system. In particular, there is a set of genes in the platypus that code for the production of proteins called b-defensins. These are small, cysteine-rich peptides that are rather like the "bullets" of the immune system; they can bind to viral coat proteins and punch holes in bacterial membranes. We humans have many epithelial cells that secrete b-defensins onto our skin and the lining of our gut and respiratory tract to kill invaders. The cells of our immune system also spew these proteins onto foreign and phagocytized cells to kill them. The platypus has repurposed the b-defensin genes, making copies that have been selected for more effective toxicity when their product proteins are injected into other animals. One especially interesting observation is that these are the same proteins used in venomous reptiles—for instance, snake venoms also contain novel forms of b-defensins. This means that animals from two distantly related groups—the lepidosaurs and the monotremes—both use b-defensin-derived venoms (Figure 2). But does this imply that the groups' last common ancestor also used these venoms?
No, it does not, and here's why: It turns out that venomous snakes and the platypus have different duplications of the b-defensin genes. So, while co-opting these genes seems to be a common strategy for evolving venoms, the details of the gene duplications reveal that platypus venom and snake venom are independently derived features. The production of venom in these animals is therefore clearly a case of convergent evolution.
References and Recommended Reading
AFP. Neither fish nor fowl: Platypus genome decoded. May 7 (2008)
Brown, S. Top billing for platypus at end of evolution tree. Nature News 453, 138 (2008) (link to article)
Warren, W. C., et al. Genome analysis of the platypus reveals unique signatures of evolution. Nature 453, 175–183 (2008) (link to article)



 Figure 1: Emergence of traits along the mammalian lineage.
Figure 1: Emergence of traits along the mammalian lineage.


























