« Prev Next »
Although the haploid human genome consists of 3 billion nucleotides, changes in even a single base pair can result in dramatic physiological malfunctions. For example, sickle-cell anemia is a disease caused by the smallest of genetic changes. Here, the alteration of a single nucleotide in the gene for the beta chain of the hemoglobin protein (the oxygen-carrying protein that makes blood red) is all it takes to turn a normal hemoglobin gene into a sickle-cell hemoglobin gene. This single nucleotide change alters only one amino acid in the protein chain, but the results are devastating.
Beta hemoglobin (beta globin) is a single chain of 147 amino acids. As previously mentioned, in sickle-cell anemia, the gene for beta globin is mutated. The resulting protein still consists of 147 amino acids, but because of the single-base mutation, the sixth amino acid in the chain is valine, rather than glutamic acid. This substitution is depicted in Table 1.
Table 1: Single-Base Mutation Associated with Sickle-Cell Anemia
| Sequence for Wild-Type Hemoglobin | ||||||||||||
| ATG | GTG | CAC | CTG | ACT | CCT | GAG | GAG | AAG | TCT | GCC | GTT | ACT |
| Start | Val | His | Leu | Thr | Pro | Glu | Glu | Lys | Ser | Ala | Val | Thr |
| Sequence for Mutant (Sickle-Cell) Hemoglobin | ||||||||||||
| ATG | GTG | CAC | CTG | ACT | CCT | GTG | GAG | AAG | TCT | GCC | GTT | ACT |
| Start | Val | His | Leu | Thr | Pro | Val | Glu | Lys | Ser | Ala | Val | Thr |
Molecules of sickle-cell hemoglobin stick to one another, forming rigid rods. These rods cause a person's red blood cells to take on a deformed, sickle-like shape, thus giving the disease its name. The rigid, misshapen blood cells do not carry oxygen well, and they also tend to clog capillaries, causing an affected person's blood supply to be cut off to various tissues, including the brain and the heart. Therefore, when an afflicted individual exerts himself or herself even slightly, he or she often experiences terrible pain, and he or she might even undergo heart attack or stroke—all because of a single nucleotide mutation (Figure 1).
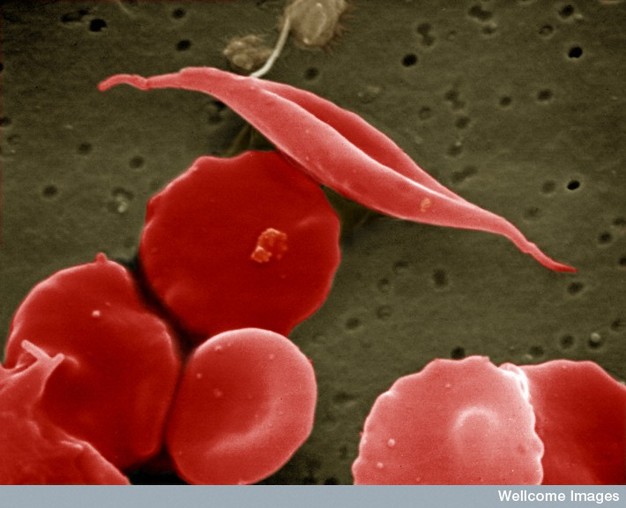
Sickle-cell anemia is one of hundreds of life-threatening disorders that are known to be caused by a change in just one of those 3 billion A's, T's, C's, or G's. Because so many diseases are associated with mutations, it is common for mutations to have a negative connotation. However, while many mutations are indeed deleterious, others are "silent"; that is, they have no discernible effect on the phenotype of an individual and remain undetected unless a molecular biologist takes a DNA sample for sequence analysis. In addition, some mutations are actually beneficial. For example, the very same mutation that causes sickle-cell anemia in affected individuals (i.e., those people who have inherited two mutant copies of the beta globin gene) can confer a survival advantage to unaffected carriers (i.e., those people who have inherited one mutant copy and one normal copy of the gene, and who generally do not show symptoms of the disease) when these people are challenged with the malaria pathogen. As a result, the sickle-cell mutation persists in populations where malaria is endemic.
Beyond the individual level, perhaps the most dramatic effect of mutation relates to its role in evolution; indeed, without mutation, evolution would not be possible. This is because mutations provide the "raw material" upon which the mechanisms of natural selection can act. By way of this process, those mutations that furnish individual organisms with characteristics better adapted to changing environmental conditions are passed on to offspring at an increased rate, thereby influencing the future of the species.
The Relationship Between Mutations and Polymorphisms
While a mutation is defined as any alteration in the DNA sequence, biologists use the term "single nucleotide polymorphism" (SNP) to refer to a single base pair alteration that is common in the population. Specifically, a polymorphism is any genetic location at which at least two different sequences are found, with each sequence present in at least 1% of the population. Note that the term "polymorphism" is generally used to refer to a normal variation, or one that does not directly cause disease. Moreover, the cutoff of at least 1% prevalence for a variation to be classified as a polymorphism is somewhat arbitrary; if the frequency is lower than this, the allele is typically regarded as a mutation (Twyman, 2003).
SNPs are important as markers, or signposts, for scientists to use when they look at populations of organisms in an attempt to find genetic changes that predispose individuals to certain traits, including disease. On average, SNPs are found every 1,000–2,000 nucleotides in the human genome, and scientists participating in the International HapMap Consortium have mapped millions of these alterations (International Human Genome Sequencing Consortium, 2001).
Types of Changes in DNA
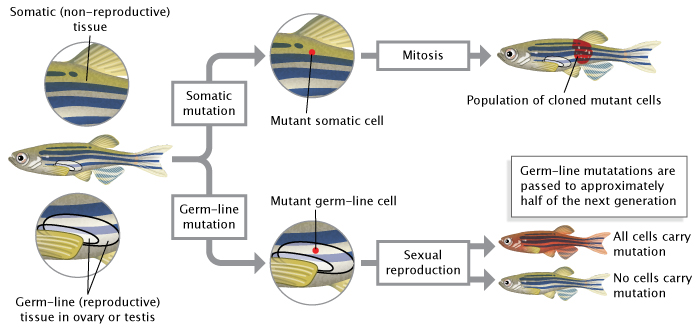
The DNA in any cell can be altered through environmental exposure to certain chemicals, ultraviolet radiation, other genetic insults, or even errors that occur during the process of replication. If a mutation occurs in a germ-line cell (one that will give rise to gametes, i.e., egg or sperm cells), then this mutation can be passed to an organism's offspring. This means that every cell in the developing embryo will carry the mutation. As opposed to germ-line mutations, somatic mutations occur in cells found elsewhere in an organism's body. Such mutations are passed to daughter cells during the process of mitosis (Figure 2), but they are not passed to offspring conceived via sexual reproduction.
As mentioned, sickle-cell anemia is the result of a change in a single nucleotide, and it represents just one class of mutations called point mutations. Changes in the DNA sequence can also occur at the level of the chromosome, in which large segments of chromosomes are altered. In this case, fragments of chromosomes can be deleted, duplicated, inverted, translocated to different chromosomes, or otherwise rearranged, resulting in changes such as modification of gene dosage, the complete absence of genes, or the alteration of gene sequence. The type of variation that occurs when entire areas of chromosomes are duplicated or lost, called copy number variation (CNV), has especially important implications for human disease and evolution. Table 2 summarizes the types of mutations and provides examples of various diseases associated with each.
| Class of Mutation | Type of Mutation | Description | Human Disease(s) Linked to This Mutation |
| Point mutation | Substitution | One base is incorrectly added during replication and replaces the pair in the corresponding position on the complementary strand | Sickle-cell anemia |
| Insertion | One or more extra nucleotides are inserted into replicating DNA, often resulting in a frameshift | One form of beta-thalassemia | |
| Deletion | One or more nucleotides is "skipped" during replication or otherwise excised, often resulting in a frameshift | Cystic fibrosis | |
| Chromosomal mutation | Inversion | One region of a chromosome is flipped and reinserted | Opitz-Kaveggia syndrome |
| Deletion | A region of a chromosome is lost, resulting in the absence of all the genes in that area | Cri du chat syndrome | |
| Duplication | A region of a chromosome is repeated, resulting in an increase in dosage from the genes in that region | Some cancers | |
| Translocation | A region from one chromosome is aberrantly attached to another chromosome | One form of leukemia | |
| Copy number variation | Gene amplification | The number of tandem copies of a locus is increased | Some breast cancers |
| Expanding trinucleotide repeat | The normal number of repeated trinucleotide sequences is expanded | Fragile X syndrome, Huntington's disease |
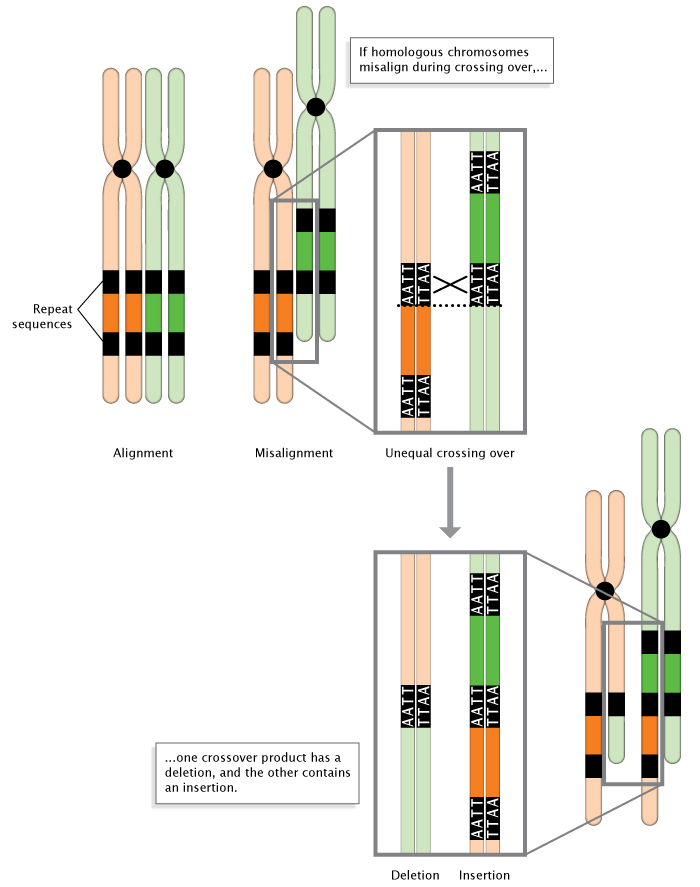
Mutations can result from a number of events, including unequal crossing-over during meiosis (Figure 3). In addition, some areas of the genome simply seem to be more prone to mutation than others. These "hot spots" are often a result of the DNA sequence itself being more accessible to mutagens. Hot spots include areas of the genome with highly repetitive sequences, such as trinucleotide repeats, in which a sequence of three nucleotides is repeated many times. During DNA replication, these repeat regions are often altered because the polymerase can "slip" as it disassociates and reassociates with the DNA strand (Viguera et al., 2001). To better understand a polymerase slip, imagine you are reading a page of text that is a repeat of a simple sequence. Say that the whole page is just copies of the word "And" ("And And And..."). Now, imagine that while reading the page, you briefly glance away and then look back at the text. It's quite likely that you will have lost your place. As a result, you may read the wrong number of copies from the page. Similarly, DNA polymerase sometimes slips and makes mistakes when reading repeats.
In other cases, mutations alter the way a gene is read through either the insertion or the deletion of a single base. In these so-called frameshift mutations, entire proteins are altered as a result of the deletion or insertion. This occurs because nucleotides are read by ribosomes in groups of three, called codons. Thus, if the number of bases removed or inserted from a gene is not a multiple of three, the reading frame for the rest of the protein is thrown off. To better understand this concept, consider the following sentence composed entirely of three-letter words, which provides an analogy for a series of three-letter codons:
THE BIG BAD FLY HAD ONE RED EYE AND ONE BLU EYE.
Now, say that a mutation eliminates the first G. As a result, the rest of the sentence is read incorrectly:
THE BIB ADF LYH ADO NER EDE YEA NDO NEB LUE YE.
The same will happen in a protein. For example, a protein might have the following coding sequence:
AUG AAA CUU CGC AGG AUG AUG AUG
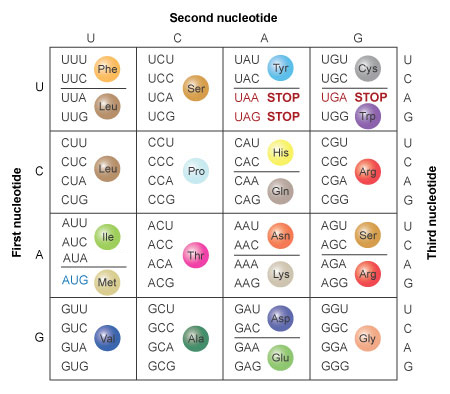
A codon translation table (Figure 4) can be used to determine that this mRNA sequence would encode the following stretch of protein:
Met-Lys-Leu-Arg-Arg-Met-Met-Met
Now, suppose that a mutation removes the fourth nucleotide. The resulting code, separated into triplet codons, would read as follows:
AUG AAC UUC GCA GGA UGA UGA UG
This would encode the following stretch of protein:
Met-Asn-Phe-Ala-Gly-STOP-STOP
Each of the STOP codons tells the ribosome to terminate protein synthesis at that point. Thus, the mutant protein is entirely different due to the deletion, and it's shorter due to the premature stop codon.
How Mutations Occur
As previously mentioned, DNA in any cell can be altered by way of a number of factors, including environmental influences, certain chemicals, spontaneous mutations, and errors that occur during the process of replication. Each of these mechanisms is discussed in greater detail in the following sections.
Mutations and the Environment
UV light can also cause covalent bonds to form between adjacent pyrimidine bases on a DNA strand, which results in the formation of pyrimidine dimers. Repair machinery exists to cope with these mutations, but it is somewhat prone to error, which means that some dimers go unrepaired. Furthermore, some people have an inherited genetic disorder called xeroderma pigmentosum (XP), which involves mutations in the genes that code for the proteins involved in repairing UV-light damage. In people with XP, exposure to UV light triggers a high frequency of mutations in skin cells, which in turn results in a high occurrence of skin cancer. As a result, such individuals are unable to go outdoors during daylight hours.
In addition to ultraviolet light, organisms are exposed to more energetic ionizing radiation in the form of cosmic rays, gamma rays, and X-rays. Ionizing radiation induces double-stranded breaks in DNA, and the resulting repair can likewise introduce mutations if carried out imperfectly. Unlike UV light, however, these forms of radiation penetrate tissue well, so they can cause mutations anywhere in the body.
Mutations Caused by Chemicals
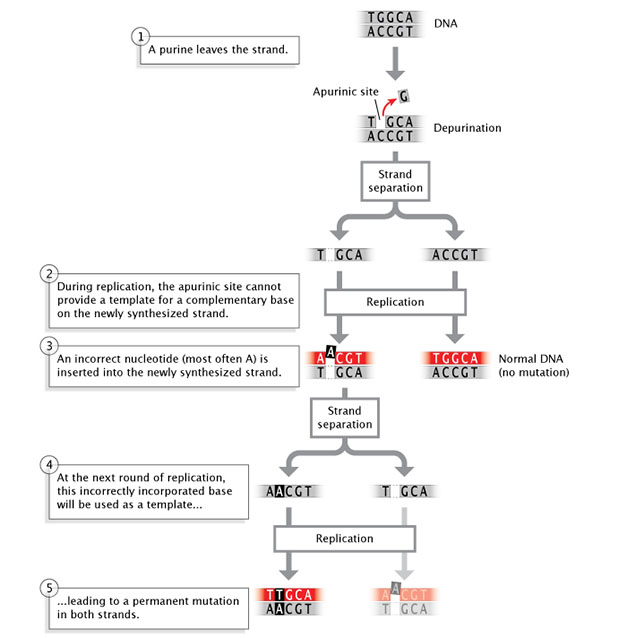
Spontaneous Mutations
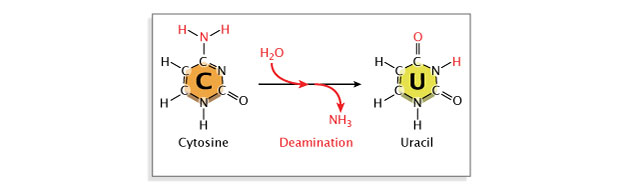
Deamination, or the removal of an amine group from a base, may also occur. Deamination of cytosine converts it to uracil, which will pair with adenine instead of guanine at the next replication, resulting in a base substitution. Repair enzymes can recognize uracil as not belonging in DNA, and they will normally repair such a lesion. However, if the cytosine residue in question is methylated (a common modification involved in gene regulation), deamination will instead result in conversion to thymine. Because thymine is a normal component of DNA, this change will go unrecognized by repair enzymes (Figure 6).
Errors During DNA Replication
Errors that occur during DNA replication play an important role in some mutations, especially trinucleotide repeat (TNR) expansions. It is thought that the ability of repeat sequences to form secondary structures, such as intrastrand hairpins, during replication might contribute to slippage of DNA polymerase, causing this enzyme to slide back and repeat replication of the previous segment (Figure 7). Supporting this hypothesis, lagging-strand synthesis has been shown to be particularly sensitive to repeat expansion. For instance, the secondary structure of some TNR DNA has been shown to inhibit an enzyme (FEN1) necessary for proper resolution of the Okazaki fragments generated during lagging-strand replication; as a result, FEN1 mutant yeast cells demonstrate increased expansion of CAG repeats.
As previously mentioned, repeats also occur in nonmitotic tissue, and CAG repeats have further been shown to accumulate in mice defective for individual DNA repair pathways, suggesting that multiple repair mechanisms must be operative in repeat expansion in nonproliferating cells (Pearson et al., 2005). In agreement with this hypothesis, studies have revealed increased repeat instability following induction of double-stranded breaks and UV-induced lesions, which are corrected by nucleotide excision repair.
To date, all diseases associated with TNRs involve repeat instability upon transmission from parent to offspring, often in a sex-specific manner. For example, the CAG repeats that characterize Huntington's disease typically exhibit greater expansion when inherited paternally. This expansion has been shown to occur prior to meiosis, when germ cells are proliferating. Contraction of other TNRs has been linked to sex-specific differences in germ-line DNA methylation patterns (Pearson et al., 2005).
Mutations, DNA Repair, and Evolution
Thus, mutations are not always a result of mutagens encountered in the environment. There is a natural—albeit low—error rate that occurs during DNA replication. In most cases, the extensive network of DNA repair machinery that exists in the cell halts cell division before an incorrectly placed nucleotide is set in place and a mismatch is made in the complementary strand. However, if the repair machinery does not catch the mistake before the complementary strand is formed, the mutation is established in the cell. This mutation can then be inherited in daughter cells or in embryos (if the mutation has occurred in the germ line).
Together, these different classes of mutations and their causes serve to place organisms at risk for disease and to provide the raw material for evolution. Thus, mutations are often detrimental to individuals, but they serve to diversify the overall population.
References and Recommended Reading
Denissenko, M. F., et al. Preferential formation of benzo[a]pyrene adducts at lung cancer mutational hotspots in P53. Science 274, 430–432 (1996)
Greenblatt, M. S., et al. Mutations in the P53 tumor suppressor gene: Clues to cancer etiology and molecular pathogenesis. Cancer Research 54, 4855–4878 (1994)
International Human Genome Sequencing Consortium. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001) (link to article)
Kimchi-Sarfaty, C., et al. "Silent" polymorphism in the MDR1 gene changes substrate specificity. Science 315, 525–528 (2006)
Mulligan, L. M., et al. Germ-line mutations of the RET proto-oncogene in multiple endocrine neoplasia type 2A. Nature 363, 458–460 (1993) (link to article)
Nells, E., et al. PMP22 Thr (118) Met: Recessive CMT1 mutation or polymorphism? Nature 15, 13–14 (1997) (link to article)
Pearson, C. E., et al. Repeat instability: Mechanisms of dynamic mutations. Nature Reviews Genetics 6, 729–742 (2005) (link to article)
Pierce, B. A. Genetics: A Conceptual Approach (Freeman, New York, 2000)
Seidl, H., et al. Ultraviolet exposure as the main initiator of P53 mutations in basal cell carcinomas from psoralen and ultraviolet A-treated patients with psoriasis. Journal of Investigative Dermatology 117, 365–370 (2001)
Twyman, R. Mutation or polymorphism? Wellcome Trust website, http://genome.wellcome.ac.uk/doc_WTD020780.html (2003)
Viguera, E., et al. Replication slippage involves DNA polymerase pausing and dissociation. EMBO Journal 20, 2587–2595 (2001)




 Figure 7
Figure 7


























