« Prev Next »
It is well known that errors during mitosis and meiosis can result in duplications and deletions of genes on a chromosomal level, which can lead to disorders. In fact, in the days prior to DNA sequencing, rare changes in gene numbers could actually be detected at the chromosomal level using a microscope. Nonetheless, scientists did not think wide-scale variations in gene copy numbers existed on a subchromosomal scale, nor did they believe that any such variations could lead to disease. Only recently did confusing laboratory results stimulate investigators to ask whether all autosomal genes are present in two copies, with a single allele inherited from each parent. By that time, the Human Genome Project and advances in genotyping provided the tools necessary to investigate variations in gene copy numbers on a subchromosomal scale (Lander et al., 2001; Venter et al., 2001).
Discovering Copy Number Variants
In 2002, Charles Lee was trying to genotype patients, but his experiments were repeatedly unsuccessful. He was finding that healthy control patients showed major variations in their genetic sequences, with some having more copies of specific genes than others. Lee began to collaborate with Steven Scherer, who had made similar observations, and together their labs used array-based comparative genomic hybridization approaches to measure the occurrence of these copy variants across the genome. Meanwhile, Michael Wigler was also observing differences in copy numbers in healthy individuals using a complementary microarray technique involving representational oligonucleotide probes to detect amplifications and deletions in the genome (Check, 2005). Thus, in 2004, both sets of researchers published findings that indicated large-scale variations in copy number were common and occurred in hundreds of places in the human genome, including areas coding for disease-related genes (Iafrate et al., 2004; Sebat et al., 2004; Figure 1).
These differences were named copy number variants, and they describe a segment of DNA that is 1 kilobase or larger and present at a variable copy number in comparison with a reference genome (Feuk et al., 2006). Copy number variants are mutations and can include deletions, insertions, and duplications. Sometimes, a copy number variant may even be so large that half a million nucleotides are affected.
Copy Number Variants and Disease

Upon learning of copy number variants, scientists immediately began to speculate that they might underlie genetic diversity and susceptibility to certain diseases, including neurological disorders and leukemia (Iafrate et al., 2004; Sebat et al., 2004). For example, after studying 270 individuals, Redon et al. (2006) discovered that copy number variants covered approximately 12% of the human genome; another research team determined that there is an average of 12 copy number variants per individual (Feuk et al., 2006). Given these values, it seems that the sheer scale of copy number variants in our genome might profoundly affect our health. Indeed, there is growing interest in the influence of this variation upon complex disease phenotypes (Beckmann et al., 2007; Figure 2) because approximately half of the copy number variants detected so far overlap with protein-coding regions (Sebat et al., 2004).
Most copy number variants exist in healthy individuals; however, these variants are hypothesized to cause diseases through several mechanisms, as shown in Figure 3. First, copy number variants can directly influence gene dosage through insertions or deletions, which can result in altered gene expression and potentially cause genetic diseases. Gene dosage describes the number of copies of a gene in a cell, and gene expression can be influenced by higher and lower gene dosages. For example, deletions can result in a lower gene dosage or copy number than what is normally expressed by removing a gene entirely (Figure 3a). Deletions can also result in the unmasking of a recessive allele that would normally not be expressed (Figure 3b). Structural variants that overlap a gene can reduce or prevent the expression of the gene through inversions, deletions, or translocations (Figure 3b). Variants can also affect a gene's expression indirectly by interacting with regulatory elements. For instance, if a regulatory element is deleted, a dosage-sensitive gene might have lower or higher expression than normal (Figure 3c). Sometimes, the combination of two or more copy number variants can produce a complex disease, whereas individually the changes produce no effect (Figure 3d). Some variants are flanked by homologous repeats, which can make genes within the copy number variant susceptible to nonallelic homologous recombination and can predispose individuals or their descendants to a disease (Freeman et al., 2006). Additionally, complex diseases might occur when copy number variants are combined with other genetic and environmental factors (Feuk et al., 2006).
For example, certain breast cancers are associated with overexpression of the ERBB2 gene, which codes for human epidermal growth factor receptor 2. Copy number variations and other types of mutations can cause overexpression (Pollack et al., 1999). Measuring high copies of ERBB2 is associated with aggressive forms of breast cancer and is a major target of treatment (Peiró, 2004). Therefore, measuring the ERBB2 copy number can provide a diagnostic tool for breast cancer and other cancers. Similarly, copy number variations were identified on chromosome 22 in regions involved with spinal muscle atrophy and DiGeorge syndrome, as well as in the imprinted chromosome 15 region associated with Prader-Willi syndrome and Angelman syndrome (Redon et al., 2006). These diseases might be caused by copy number variants due to inversions and deletions in critical genes. Copy number variants were also detected in genetic regions associated with complex neurological diseases, such as Alzheimer's disease and schizophrenia (Freeman et al., 2006; Redon et al., 2006).
By using DNA microarrays, scientists are now screening patients with genetic diseases and comparing them to unaffected control individuals to examine which copy number variants are truly associated with disease states and which are common in the population. Information about these copy number variants might allow for the identification of specific disease-related genes that were previously unknown.
Copy Number Variants in Healthy Individuals
As previously mentioned, copy number variants do not necessarily have a negative effect on health. Consider, for example, chemokine CCL3L1, which can potently suppress human immunodeficiency virus 1 (HIV-1). Researcher Enrique Gonzalez and his colleagues found that individuals who carried fewer copy number variants encoding CCL3L1 than average were significantly more susceptible to HIV and acquired immunodeficiency syndrome (AIDS) (Gonzalez et al., 2005). This means that bearing extra copies of CCL3L1 can protect an individual against contracting HIV and developing AIDS. Similarly, other copy number variants carried by healthy individuals that seem to have no function might actually be evolutionarily retained in populations if they provide a selective advantage.
Current research aims to identify the functional mechanisms by which copy number variation cause diseases. Although preliminary findings suggest that the presence of copy number variants might be associated with certain disease phenotypes, the variants are not necessarily the causes of these diseases (McCarroll & Altshuler, 1999). As studies relating copy number variation to diseases expand, our understanding of human diversity, the causes and development of complex diseases, and disease resistance will grow accordingly, which will allow the development of improved diagnostic and treatment strategies.
References and Recommended Reading
Beckmann, J. S., et al. Copy number variants and genetic traits: Closer to the resolution of phenotypic to genotypic variability. Nature Reviews Genetics 8, 639–646 (2007) doi:10.1038/nrg2149 (link to article)
Check, E. Patchwork people. Nature 437, 1084–1086 (2005) doi:10.1038/4371084a (link to article)
Feuk, L., et al. Structural variation in the human genome. Nature Reviews Genetics 7, 85–97 (2006) doi:10.1038/nrg1767 (link to article)
Freeman, J. L., et al. Copy number variation: New insights in genome diversity. Genome Research 16, 949–961 (2006) doi:10.1101/gr.3677206
Gonzalez, E., et al. The influence of CCL3L1 gene-containing segmental duplications on HIV-1/AIDS susceptibility. Science 307, 1434 (2005) doi:10.1126/science.1101160
Iafrate, A. J., et al. Detection of large-scale variation in the human genome. Nature Genetics 36, 949–951 (2004) doi:10.1038/ng1416 (link to article)
Lander, E. S., et al. Initial sequencing and analysis of the human genome. Nature 409, 860–921 (2001) (link to article)
McCarroll, S. A., & Altshuler, D. M. Copy number variation and association studies of human disease. Nature Genetics 39, S37–42 (2007) doi:10.1038/ng2080 (link to article)
Peiró, G., et al. Analysis of HER-2/neu amplification in endometrial carcinoma by chromogenic in situ hybridization. Correlation with fluorescence in situ hybridization, HER-2/neu, p53 and Ki-67 protein expression, and outcome. Modern Pathology 17, 277–287 (2004) doi:10.1038/modpathol.3800006
Pollack, J. R., et al. Genome-wide analysis of DNA copy number changes using cDNA microarrays. Nature Genetics 23, 41–46 (1999) doi:10.1038/12640 (link to article)
Redon, R., et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006) (link to article)
Sebat, J., et al. Large-scale copy number polymorphism in the human genome. Science 305, 525–528 (2004)
Venter, J. C., et al. The sequence of the human genome. Science 291, 1304–1351(2001)



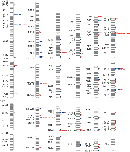
 Figure 1
Figure 1