« Prev Next »
The postulated 99.9% genetic identicalness of all humans has been recently called into question due to an improved understanding of structural variations in human DNA. While most initial studies of genetic variation concentrated on individual nucleotide sequences, investigators have also found that large-scale changes occur in many locations throughout the genome. These insertions, deletions, inversions, and duplications result in changes in the physical arrangement of genes on chromosomes. Indeed, for as long as cytogeneticists have studied chromosomes under microscopes, they have observed variations in chromosomal structure. These scientists have noted such anomalies as aneuploidy (abnormal chromosome number), translocations of material from one chromosome to another, large-scale deletions and insertions, fragile sites, and variations in the size of the Y chromosome (Feuk et al., 2006).
The duplication of the Bar gene in Drosophila was one of the earliest structural variations to be linked to a phenotype. Seventy years ago, this variation was shown to cause the eye field of affected flies to be much narrower than that of flies with wild-type eyes (Bridges, 1936). In yet another example of a phenotypic link to a chromosomal anomaly, in humans, the duplication of part or all of chromosome 21 has been associated with Down syndrome. This duplication may be the result of nondisjunction or of translocation.
More recently, both aneuploidy and chromosomal translocations have been causally implicated in human cancers. These changes usually arise in individual somatic cells. While the study of such structural variations was initially limited to individual changes that could be seen through light microscopes, the advent of new technologies has allowed identification of submicroscopic structural variations on a genome-wide scale.
Discovering and Defining CNVs
CNVs have been found in all human populations, as well as in other mammalian species (Freeman et al., 2006). Perhaps the best-defined and most widely known CNVs are the trinucleotide repeats (TNRs), which consist of three nucleotides repeating in tandem. TNRs exhibit dynamic expansion and contraction in a number of disease states, such as fragile X syndrome and Huntington's disease, with the number of repeats varying in both normal and afflicted individuals. In most cases, TNRs exhibit expansion with age. Because they are inherited through families, increased copy numbers typically correlate with greater disease severity and/or earlier onset of symptoms. Contraction of TNRs has been observed less frequently than expansion, typically upon paternal inheritance (Pearson et al., 2005).
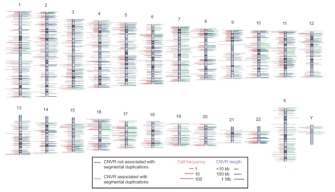
Recently, a collaboration of international research laboratories has begun compiling a complete catalog of existing CNVs in the human genome. An examination of 270 DNA samples from the multiethnic population employed by the HapMap Project revealed a total of 1,447 discrete CNVs. Taken together, these CNVs cover approximately 360 megabases, or 12% of the human genome. The HapMap Project notes that CNVs encompass more nucleotide content per genome than SNPs, underscoring CNVs' significance to genetic diversity. A genome-wide map of CNVs shows that no region of the genome is exempt, and that the percentage of an individual's chromosomes that exhibit CNVs varies anywhere from 6% to 19% (Figure 1; Redon et al., 2006).
CNVs and Evolution
Analysis of copy number variation in the human and chimpanzee genomes demonstrates the potentially greater role of CNVs in evolutionary change than single base-pair sequence variation (Cheng et al., 2005). Comparisons of the human and chimpanzee genomes revealed that there are more than twice as many nucleotides involved in CNVs as there are in changes to individual nucleotides, 2.7% compared to 1.2%. Furthermore, research by Cheng et al. (2005) revealed that the majority of CNVs were shared between the human and chimpanzee genomes, but approximately one-third of the CNVs observed in the human genome were unique to our species. In many cases, other researchers were able to confirm these results, based on a comparison of genomic sequences with comparative genomic hybridization. Additional studies have further revealed that CNVs are often linked to genetic diseases apparent in humans (Stankiewicz & Lupski, 2002).
Yet another study (Stefansson et al., 2005) identified an inversion currently undergoing positive selection in humans. This variation involves an inversion of 900 kb, flanked by duplicated sequences that vary widely in copy number. The study, which analyzed tens of thousands of samples, showed that females who carried the inversion had more children than noncarriers. The haplotypes of carriers and noncarriers of the inversion were then examined for diversity in different populations. Simulations of coalescence suggested that the frequency of the inversion did not arise from neutral evolution, though the mechanism by which this polymorphism confers a selective advantage remains unknown.
Redon et al. (2006) also addressed the potential role of CNVs in evolution by examining the persistence of distinct categories of structural changes through multiple generations. These researchers proposed that, while deletions are typically selected against, the existence of gene families indicates that duplications of genetic material can experience positive selection.
The globin genes are a prime example of how a portion of the genetic sequence can be duplicated and the resulting duplicates can gain a novel function. The beta-globin gene cluster represents several different genes that are differentially expressed at different times in development (Makova & Li, 2003). This suggests that, just like other types of mutations, structural variations can provide raw material for evolution in the form of extra genes that are free to acquire new, and potentially advantageous, functions.
Redon and colleagues (2006) specifically investigated a potential role for CNVs in evolution by examining the character of the associated DNA, including the functional categories of genes that most frequently exhibit this class of structural variant. The researchers found that CNVs typically lie outside of coding sequences and ultraconserved regulatory elements. These elements, originally identified by Bejerano and colleagues (2004), are sequences of at least 200 base pairs that are 100% conserved across several species, including humans, rats, and mice. This research also revealed that the functional categories with the greatest enrichment for CNVs were those genes involved in cell adhesion, the sensory perception of smell, and responses to chemical stimuli. These findings were in agreement with previous studies on CNV distribution (Nguyen et al., 2006). In addition, categories that were underrepresented with regard to CNV distribution included genes involved in cell signaling and cell proliferation, as well as those involved in regulation of protein phosphorylation. The researchers suggest that this discovery reflects the sensitivity of the function of these gene products to dosage effects, particularly during embryonic development, as well as the potential of these genes as oncogenes or tumor suppressor genes.
CNVs Help Differentiate Genomes
Because the study of copy number variations is a relatively new area of genetic research, many questions regarding CNVs remain unresolved. Scientists worldwide are actively pursuing research regarding the origin of these structural variations, as well as their contributions to both evolutionary adaptation and human disease. New tools, such as comparative genomic hybridization, should allow scientists to look at CNVs in detail and examine their origin and significance.
When investigators examined the raw sequence data of the human and chimpanzee genomes and estimated that there is 99.9% identicalness between the two species, they focused primarily on differences at the level of single nucleotide morphisms. Recent analysis of the structural level — specifically CNVs — has revealed an additional source of variation, and this has led to a revised picture of genomic diversity and a greater appreciation of its dynamic nature.
References and Recommended Reading
Bejerano, G., et al. Ultraconserved elements in the human genome. Science 304, 1321–1325 (2004) doi:10.1126/science.1098119
Bridges, C. B. The Bar "gene": A duplication. Science 83, 210–211 (1936) doi:10.1126/science.83.2148.210
Cheng, Z., et al. A genome-wide comparison of recent chimpanzee and human segmental duplications. Nature 437, 88–93 (2005) doi:10.1038/nature04000 (link to article)
Eichler, E. E., et al. Completing the map of human genetic variation. Nature 447, 161–165 (2007) doi:10.1038/447161a (link to article)
Feuk, L., et al. Structural variation in the human genome. Nature Reviews Genetics 7, 85–97 (2006) doi:10.1038/nrg1767 (link to article)
Freeman, J. L., et al. Copy number variation: New insights in genome diversity. Genome Research 16, 949–961 (2006)
Lakich, D., et al. Inversions disrupting the factor VIII gene are a common cause of severe haemophilia. Nature Genetics 5, 236–241 (1993) doi:10.1038/ng1193-236 (link to article)
Makova, K. D, & Li, W. H. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Research 13, 1638–1645 (2003)
Nguyen, D. Q., et al. Bias of selection on human copy number variants. PLoS Genetics 2, e2 (2006) (link to article)
Pearson, C. E., et al. Repeat instability: Mechanisms of dynamic mutations. Nature Reviews Genetics 6, 729–742 (2005) doi:10.1038/nrg1689 (link to article)
Redon, R., et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006) doi:10.1038/nature05329 (link to article)
Stankiewicz, P., & Lupski, J. R. Genomic architecture, rearrangements, and genomic disorders. Trends in Genetics 18, 74–82 (2002)
Stefansson, H., et al. A common inversion under selection in the human genome. Nature Genetics 37, 129–137 (2005) doi:10.1038/ng1508 (link to article).


 Figure 1: Genomic distribution of copy number variable regions (CNVRs).
Figure 1: Genomic distribution of copy number variable regions (CNVRs).


























