Abstract
The potential roles of epigenetic alterations in the pathogenesis of autoimmune rheumatic diseases are raising great expectations among clinicians and researchers. Epigenetic mechanisms regulate gene expression and are sensitive to external stimuli, bridging the gap between environmental and genetic factors. Considerable evidence of epigenetic changes, particularly altered patterns of DNA methylation, exists in diseases such as systemic lupus erythematosus (SLE) and rheumatoid arthritis. The importance of such changes in the pathology of rheumatic diseases has been demonstrated by examining the relationship between gene-specific methylation and SLE in monozygotic twins discordant for the disease, in whom genetic variability is excluded as a cause for discordance. Several studies have highlighted the importance of the tissue-specificity of DNA methylation changes, an aspect which—in contrast with genetic analysis—must be considered when designing epigenetic studies. Here I discuss the proposed mechanisms and implications of DNA methylation changes in the pathogenesis of autoimmune rheumatic diseases, the prospects for future epigenetic studies in rheumatology, the relevance of specific DNA methylation markers and the potential use of drugs with an epigenetic effect in the clinical management of these diseases.
Key Points
-
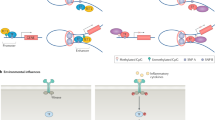
Autoimmune rheumatic disorders are complex diseases that involve genetic and environmental components—these facets are linked by epigenetic modifications, which control gene expression and are subject to environmental influences
-
Monozygotic twins discordant for autoimmune disease provide an opportunity to specifically study epigenetic changes that lead to the development of autoimmunity, because genetic variability is excluded
-
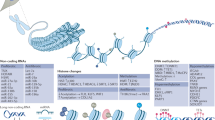
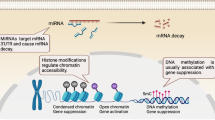
Candidate gene approaches have identified a small set of genes that undergo aberrant DNA demethylation and overexpression in systemic lupus erythematosus and rheumatoid arthritis
-
High-throughput approaches are necessary for screening epigenetic alterations in autoimmune disease, and it is essential to screen the specific tissue and cell types that are relevant to disease pathogenesis
-
Identification of cell-specific targets of epigenetic deregulation in autoimmune rheumatic disorders will provide clinical markers for diagnosis, disease progression and response to therapies
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Lie, B. A. & Thorsby, E. Several genes in the extended human MHC contribute to predisposition to autoimmune diseases. Curr. Opin. Immunol. 17, 526–531 (2005).
Delgado-Vega, A., Sánchez, E., Löfgren, S., Castillejo-López, C. & Alarcón-Riquelme, M. E. Recent findings on genetics of systemic autoimmune diseases. Curr. Opin. Immunol. 22, 698–705 (2010).
Niewold, T. B. et al. Association of the IRF5 risk haplotype with high serum interferon-α activity in systemic lupus erythematosus patients. Arthritis Rheum. 58, 2481–2487 (2008).
Kariuki, S. N. et al. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-α in lupus patients in vivo. J. Immunol. 182, 34–38 (2009).
Gourley, M. & Miller, F. W. Mechanisms of disease: Environmental factors in the pathogenesis of rheumatic disease. Nat. Clin. Pract. Rheumatol. 3, 172–180 (2007).
Klareskog, L., Padyukov, L., Lorentzen, J. & Alfredsson, L. Mechanisms of disease: genetic susceptibility and environmental triggers in the development of rheumatoid arthritis. Nat. Clin. Pract. Rheumatol. 2, 425–433 (2006).
Esteller, M. Epigenetics in cancer. N. Engl. J. Med. 358, 1148–1159 (2008).
Allis, C. D., Jenuwein, T. & Reinberg, D. in Epigenetics Ch. 3: Overview and Concepts (eds Allis, C. D., Jenuwein, T. & Reinberg, D.) 23–62 (Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, 2007).
Huertas, D., Sendra, R. & Muñoz, P. Chromatin dynamics coupled to DNA repair. Epigenetics 4, 31–42 (2009).
Corpet, A. & Almouzni, G. Making copies of chromatin: the challenge of nucleosomal organization and epigenetic information. Trends Cell Biol. 19, 29–41 (2009).
Bird, A. Perceptions of epigenetics. Nature 447, 396–398 (2007).
Sandelin, A. et al. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 8, 424–436 (2007).
Irizarry, R. A. et al. The human colon cancer methylome shows similar hypo- and hypermethylation at conserved tissue-specific CpG island shores. Nat. Genet. 41, 178–186 (2009).
Goll, M. G. & Bestor, T. H. Eukaryotic cytosine methyltransferases. Annu. Rev. Biochem. 74, 481–514 (2005).
Garcia-Manero, G. Demethylating agents in myeloid malignancies. Curr. Opin. Oncol. 20, 705–710 (2008).
Bhutani, N. et al. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature 463, 1042–1047 (2010).
Bruniquel, D. & Schwartz, R. H. Selective, stable demethylation of the interleukin-2 gene enhances transcription by an active process. Nat. Immunol. 4, 235–240 (2003).
Ooi, S. K. & Bestor, T. H. The colorful history of active DNA demethylation. Cell 133, 1145–1148 (2008).
Fritz, E. L. & Papavasiliou, F. N. Cytidine deaminases: AIDing DNA demethylation? Genes Dev. 24, 2107–2114 (2010).
Rai, K. et al. DNA demethylation in zebrafish involves the coupling of a deaminase, a glycosylase, and gadd45. Cell 135, 1201–1212 (2008).
Salvador, J. M. et al. Mice lacking the p53-effector gene Gadd45a develop a lupus-like syndrome. Immunity 16, 499–508 (2002).
Tahiliani, M. et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science 324, 930–935 (2009).
Ito, S. et al. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature 466, 1129–1133 (2010).
Kouzarides, T. Chromatin modifications and their function. Cell 128, 693–705 (2007).
Allis, C. D. et al. New nomenclature for chromatin-modifying enzymes. Cell 131, 633–636 (2007).
Biancotto, C., Frigè, G. & Minucci, S. Histone modification therapy of cancer. Adv. Genet. 70, 341–386 (2010).
Kraus, W. L. & Wong, J. Nuclear receptor-dependent transcription with chromatin. Is it all about enzymes? Eur. J. Biochem. 269, 2275–2283 (2002).
Schwartz, Y. B. et al. Alternative epigenetic chromatin states of polycomb target genes. PLoS Genet. 6, e1000805 (2010).
Bell, C. G. & Beck, S. The epigenomic interface between genome and environment in common complex diseases. Brief. Funct. Genomics 9, 477–485 (2010).
Richly, H., Lange. M., Simboeck. E. & Di Croce, L. Setting and resetting of epigenetic marks in malignant transformation and development. Bioessays 32, 669–679 (2010).
Esteller, M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat. Rev. Genet. 8, 286–298 (2007).
Ehrlich, M. DNA hypomethylation in cancer cells. Epigenomics 1, 239–259 (2009).
Konkel, M. K. & Batzer, M. A. A mobile threat to genome stability: The impact of non-LTR retrotransposons upon the human genome. Semin. Cancer Biol. 20, 211–221 (2010).
Schlesinger, Y. et al. Polycomb-mediated methylation on Lys27 of histone H3 pre-marks genes for de novo methylation in cancer. Nat. Genet. 39, 232–236 (2007).
Widschwendter, M. et al. Epigenetic stem cell signature in cancer. Nat. Genet. 39, 157–158 (2007).
Di Croce, L. Chromatin modifying activity of leukaemia associated fusion proteins. Hum. Mol. Genet. 14 (Suppl. 1), R77–R84 (2005).
Ballestar, E. et al. Methyl-CpG binding proteins identify novel sites of epigenetic inactivation in human cancer. EMBO J. 22, 6335–6345 (2003).
Cooper, G. S., Miller, F. W. & Pandey, J. P. The role of genetic factors in autoimmune disease: implications for environmental research. Environ. Health Perspect. 107 (Suppl. 5), 693–700 (1999).
Salvetti, M., Ristori, G., Bomprezzi, R., Pozzilli, P. & Leslie, R. D. Twins: mirrors of the immune system. Immunol. Today 21, 342–347 (2000).
Fraga, M. F. et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA 102, 10604–10609 (2005).
Kaminsky, Z. A. et al. DNA methylation profiles in monozygotic and dizygotic twins. Nat. Genet. 41, 240–245 (2009).
Cezar, G. G. et al. Genome-wide epigenetic alterations in cloned bovine fetuses. Biol. Reprod. 68, 1009–1014 (2003).
Young, L. E. et al. Conservation of IGF2-H19 and IGF2R imprinting in sheep: effects of somatic cell nuclear transfer. Mech. Dev. 120, 1433–1442 (2003).
Heijmans, B. T. et al. Persistent epigenetic differences associated with prenatal exposure to famine in humans. Proc. Natl Acad. Sci. USA 105, 17046–17049 (2008).
Sinclair, K. D. et al. DNA methylation, insulin resistance, and blood pressure in offspring determined by maternal periconceptional B vitamin and methionine status. Proc. Natl Acad. Sci. USA 104, 19351–19356 (2007).
Cannat, A. & Seligmann, M. Induction by isoniazid and hydrallazine of antinuclear factors in mice. Clin. Exp. Immunol. 3, 99–105 (1968).
Richardson, B. Effect of an inhibitor of DNA methylation on T cells. II. 5-Azacytidine induces self-reactivity in antigen-specific T4+ cells. Hum. Immunol. 17, 456–470 (1986).
Cornacchia, E. et al. Hydralazine and procainamide inhibit T cell DNA methylation and induce autoreactivity. J. Immunol. 140, 2197–2200 (1988).
Quddus, J. et al. Treating activated CD4+ T cells with either of two distinct DNA methyltransferase inhibitors, 5-azacytidine or procainamide, is sufficient to cause a lupus-like disease in syngeneic mice. J. Clin. Invest. 92, 38–53 (1993).
Yung, R. L., Quddus, J., Chrisp, C. E., Johnson, K. J. & Richardson, B. C. Mechanism of drug-induced lupus. I. Cloned Th2 cells modified with DNA methylation inhibitors in vitro cause autoimmunity in vivo. J. Immunol. 154, 3025–3035 (1995).
Blomgren, S. E., Condemi, J. J. & Vaughan, J. H. Procainamide-induced lupus erythematosus. Clinical and laboratory observations. Am. J. Med. 52, 338–348 (1972).
Richardson, B. C. et al. Phenotypic and functional similarities between 5-azacytidine-treated T cells and a T cell subset in patients with active systemic lupus erythematosus. Arthritis Rheum. 35, 647–662 (1992).
Richardson, B. et al. Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 33, 1665–1673 (1990).
Corvetta, A., Della Bitta, R., Luchetti, M. M. & Pomponio, G. 5-Methylcytosine content of DNA in blood, synovial mononuclear cells and synovial tissue from patients affected by autoimmune rheumatic diseases. J. Chromatogr. 566, 481–491 (1991).
Huck, S. & Zouali, M. DNA methylation: a potential pathway to abnormal autoreactive lupus B cells. Clin. Immunol. Immunopathol. 80, 1–8 (1996).
Javierre, B. M. et al. Changes in the pattern of DNA methylation associated with twin discordance in systemic lupus erythematosus. Genome Res. 20, 170–179 (2010).
Fraga, M. F. & Esteller, M. Epigenetics and aging: the targets and the marks. Trends Genet. 23, 413–418 (2007).
Maegawa, S. et al. Widespread and tissue specific age-related DNA methylation changes in mice. Genome Res. 20, 332–340 (2010).
Wilson, V. L. & Jones, P. A. DNA methylation decreases in aging but not in immortal cells. Science 220, 1055–1057 (1983).
Zhang, W. et al. Comparison of global DNA methylation profiles in replicative versus premature senescence. Life Sci. 83, 475–480 (2008).
Deng, C. et al. Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 44, 397–407 (2001).
Sawalha, A. H. et al. Defective T-cell ERK signaling induces interferon-regulated gene expression and overexpression of methylation-sensitive genes similar to lupus patients. Genes Immun. 9, 368–378 (2008).
Tili, E., Michaille, J. J., Costinean, S., Croce, C. M. MicroRNAs, the immune system and rheumatic disease. Nat. Clin. Pract. Rheumatol. 4, 534–541 (2008).
Pan, W. et al. MicroRNA-21 and microRNA-148a contribute to DNA hypomethylation in lupus CD4+ T cells by directly and indirectly targeting DNA methyltransferase 1. J. Immunol. 184, 6773–6781 (2010).
Li, Y. et al. Overexpression of the growth arrest and DNA damage-induced 45α gene contributes to autoimmunity by promoting DNA demethylation in lupus T cells. Arthritis Rheum. 62, 1438–1447 (2010).
Hsu, H.-C. et al. Inhibition of the catalytic function of activation-induced cytidine deaminase (AICDA) promotes apoptosis of germinal center B cells. Arthritis Rheum. doi:10.1002/art.30257.
Lu, Q. et al. Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 46, 1282–1291 (2002).
Oelke, K. et al. Overexpression of CD70 and overstimulation of IgG synthesis by lupus T cells and T cells treated with DNA methylation inhibitors. Arthritis Rheum. 50, 1850–1860 (2004).
Lu, Q. et al. Demethylation of CD40LG on the inactive X in T cells from women with lupus. J. Immunol. 179, 6352–6358 (2007).
Kaplan, M. J., Lu, Q., Wu, A., Attwood, J. & Richardson, B. Demethylation of promoter regulatory elements contributes to perforin overexpression in CD4+ lupus T cells. J. Immunol. 172, 3652–3661 (2004).
Garaud, S. et al. IL-6 modulates CD5 expression in B cells from patients with lupus by regulating DNA methylation. J. Immunol. 182, 5623–5632 (2009).
Nile, C. J., Read, R. C., Akil, M., Duff, G. W. & Wilson, A. G. Methylation status of a single CpG site in the IL-6 promoter is related to IL-6 messenger RNA levels and rheumatoid arthritis. Arthritis Rheum. 58, 2686–2693 (2008).
Neidhart, M. et al. Retrotransposable L1 elements expressed in rheumatoid arthritis synovial tissue: association with genomic DNA hypomethylation and influence on gene expression. Arthritis Rheum. 43, 2634–2647 (2000).
Karouzakis, E., Gay, R. E., Michel, B. A., Gay, S. & Neidhart, M. DNA hypomethylation in rheumatoid arthritis synovial fibroblasts. Arthritis Rheum. 60, 3613–3622 (2009).
Stanczyk, J. et al. Altered expression of miR-203 in rheumatoid arthritis synovial fibroblasts and its role in fibroblast activation. Arthritis Rheum. 63, 373–381 (2011).
Lane, A. A. & Chabner, B. A. Histone deacetylase inhibitors in cancer therapy. J. Clin. Oncol. 27, 5459–5468 (2009).
Mishra, N. et al. Histone deacetylase inhibitors modulate renal disease in the MRL-lpr/lpr mouse. J. Clin. Invest. 111, 539–552 (2003).
Reilly, C. M. et al. Modulation of renal disease in MRL/lpr mice by suberoylanilide hydroxamic acid. J. Immunol. 173, 4171–4178 (2004).
Vojinovic, J. et al. Safety and efficacy of an oral histone deacetylase inhibitor in systemic onset juvenile idiopathic arthritis. Arthritis Rheum. doi:10.1002/art.30238.
Dörner, T., Radbruch, A. & Burmester, G. R. B-cell-directed therapies for autoimmune disease. Nat. Rev. Rheumatol. 5, 433–441 (2009).
Petri, M. et al., Association of plasma B lymphocyte stimulator levels and disease activity in systemic lupus erythematosus. Arthritis Rheum. 58, 2453–2459 (2008).
Sanz, I. & Lee, F. E. B cells as therapeutic targets in SLE. Nat. Rev. Rheumatol. 6, 326–337 (2010).
Mazari, L., Ouarzane, M. & Zouali, M. Subversion of B lymphocyte tolerance by hydralazine, a potential mechanism for drug-induced lupus. Proc. Natl Acad. Sci. USA 104, 6317–6322 (2007).
Baranzini, S. E. et al. Genome, epigenome and RNA sequences of monozygotic twins discordant for multiple sclerosis. Nature 464, 1351–1356 (2010).
Mastronardi, F. G., Noor, A., Wood, D. D., Paton, T. & Moscarello, M. A. Peptidyl argininedeiminase 2 CpG island in multiple sclerosis white matter is hypomethylated. J. Neurosci. Res. 85, 2006–2016 (2007).
Basu, D. et al. Stimulatory and inhibitory killer Ig-like receptor molecules are expressed and functional on lupus T cells. J. Immunol. 183, 3481–3487 (2009).
Mikkelsen, T. S. et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560 (2007).
Ballestar, E. & Wolffe, A. P. Methyl-CpG-binding proteins. Targeting specific gene repression. Eur. J. Biochem. 268, 1–6 (2001).
Dhasarathy, A. & Wade, P. A. The MBD protein family—reading an epigenetic mark? Mutat. Res. 647, 39–43 (2008).
Ooi, S. K. et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 448, 714–717 (2007).
de la Cruz, X., Lois, S., Sánchez-Molina, S. & Martínez-Balbás, M. A. Do protein motifs read the histone code? Bioessays 27, 164–175 (2005).
Acknowledgements
E. Ballestar is supported by PI081346 (FIS) grant from the Spanish Ministry of Science and Innovation (MICINN) and 2009SGR184 grant from AGAUR (Catalan Government).
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Rights and permissions
About this article
Cite this article
Ballestar, E. Epigenetic alterations in autoimmune rheumatic diseases. Nat Rev Rheumatol 7, 263–271 (2011). https://doi.org/10.1038/nrrheum.2011.16
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrrheum.2011.16
This article is cited by
-
Epigenetic and metabolic reprogramming in inflammatory bowel diseases: diagnostic and prognostic biomarkers in colorectal cancer
Cancer Cell International (2023)
-
Current trends in epigenetic, cellular and molecular pathways in management of rheumatoid arthritis
Inflammopharmacology (2023)
-
Clinical research progress of novel biologics for the treatment of lupus nephritis
Clinical and Experimental Medicine (2023)
-
Epigenetics in Non-tumor Immune-Mediated Skin Diseases
Molecular Diagnosis & Therapy (2021)
-
Emerging epigenetic targets in rheumatoid arthritis
Rheumatology International (2021)