Abstract
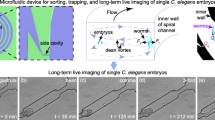
Here we describe a protocol for the fabrication and use of a microfluidic device to rapidly orient >700 Drosophila embryos in parallel for end-on imaging. The protocol describes master microfabrication (∼1 d), polydimethylsiloxane molding (few hours), system setup and device operation (few minutes) and imaging (depending on application). Our microfluidics-based approach described here is one of the first to facilitate rapid orientation for end-on imaging, and it is a major breakthrough for quantitative studies on Drosophila embryogenesis. The operating principle of the embryo trap is based on passive hydrodynamics, and it does not require direct manipulation of embryos by the user; biologists following the protocol should be able to repeat these procedures. The compact design and fabrication materials used allow the device to be used with traditional microscopy setups and do not require specialized fixtures. Furthermore, with slight modification, this array can be applied to the handling of other model organisms and oblong objects.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Bier, E. Drosophila, the golden bug, emerges as a tool for human genetics. Nat. Rev. Genet. 6, 9–23 (2005).
Adams, M.D. et al. The genome sequence of Drosophila melanogaster. Science 287, 2185–2195 (2000).
Gurdon, J.B. & Bourillot, P.Y. Morphogen gradient interpretation. Nature 413, 797–803 (2001).
Frohnhofer, H.G. & Nusslein-Volhard, C. Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature 324, 120–125 (1986).
Roth, S., Stein, D. & Nusslein-Volhard, C. A gradient of nuclear localization of the dorsal protein determines dorsoventral pattern in the Drosophila embryo. Cell 59, 1189–1202 (1989).
Steward, R. Relocalization of the Dorsal protein from the cytoplasm to the nucleus correlates with its function. Cell 59, 1179–1188 (1989).
Rushlow, C.A. & Shvartsman, S.Y. Temporal dynamics, spatial range, and transcriptional interpretation of the Dorsal morphogen gradient. Curr. Opin. Genet. Dev. 22, 542–546 (2012).
Chung, K. et al. A microfluidic array for large-scale ordering and orientation of embryos. Nat. Methods 8, 171–176 (2011).
Kanodia, J.S. et al. A computational statistics approach for estimating the spatial range of morphogen gradients. Development 138, 4867–4874 (2011).
Kanodia, J.S. et al. Pattern formation by graded and uniform signals in the early Drosophila embryo. Biophys. J. 102, 427–433 (2012).
Helman, A. et al. RTK signaling modulates the Dorsal gradient. Development 139, 3032–3039 (2012).
Kim, Y. et al. Gene regulation by MAPK substrate competition. Dev. Cell 20, 880–887 (2011).
Crane, M.M., Chung, K. & Lu, H. Computer-enhanced high-throughput genetic screens of C. elegans in a microfluidic system. Lab Chip 9, 38–40 (2009).
Chung, K., Crane, M.M. & Lu, H. Automated on-chip rapid microscopy, phenotyping and sorting of C. elegans. Nat. Methods 5, 637–643 (2008).
Crane, M.M., Chung, K., Stirman, J. & Lu, H. Microfluidics-enabled phenotyping, imaging, and screening of multicellular organisms. Lab Chip 10, 1509–1517 (2010).
Liang, H.L. et al. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature 456, 400–403 (2008).
Li, X.Y. et al. Transcription factors bind thousands of active and inactive regions in the Drosophila blastoderm. PLoS Biol. 6, e27 (2008).
Reeves, G.T. et al. Dorsal-ventral gene expression in the Drosophila embryo reflects the dynamics and precision of the dorsal nuclear gradient. Dev. Cell 22, 544–557 (2012).
Chung, K.H., Rivet, C.A., Kemp, M.L. & Lu, H. Imaging single-cell signaling dynamics with a deterministic high-density single-cell trap array. Anal. Chem. 83, 7044–7052 (2011).
Akagi, J. et al. Miniaturized embryo array for automated trapping, immobilization and microperfusion of zebrafish embryos. PLoS ONE 7, 36630 (2012).
Kohli, V. et al. An alternative method for delivering exogenous material into developing zebrafish embryos. Biotechnol. Bioeng. 98, 1230–1241 (2007).
Samara, C. et al. Large-scale in vivo femtosecond laser neurosurgery screen reveals small-molecule enhancer of regeneration. Proc. Natl Acad. Sci. USA 107, 18342–18347 (2010).
Tomer, R., Khairy, K., Amat, F. & Keller, P.J. Quantitative high-speed imaging of entire developing embryos with simultaneous multiview light-sheet microscopy. Nat. Methods 9, 755–U181 (2012).
Keller, P.J., Schmidt, A.D., Wittbrodt, J. & Stelzer, E.H.K. Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065–1069 (2008).
Fakhoury, J.R., Sisson, J.C. & Zhang, X. Microsystems for controlled genetic perturbation of live Drosophila embryos: RNA interference, development robustness and drug screening. Microfluid. Nanofluid. 6, 299–313 (2009).
Haywood, A.F. & Staveley, B.E. Parkin counteracts symptoms in a Drosophila model of Parkinson's disease. BMC Neurosci. 5, 14 (2004).
Liberman, L.M., Reeves, G.T. & Stathopoulos, A. Quantitative imaging of the Dorsal nuclear gradient reveals limitations to threshold-dependent patterning in Drosophila. Proc. Natl Acad. Sci. USA 106, 22317–22322 (2009).
Kanodia, J.S. et al. Dynamics of the Dorsal morphogen gradient. Proc. Natl Acad. Sci. USA 106, 21707–21712 (2009).
Witzberger, M.M., Fitzpatrick, J.A., Crowley, J.C. & Minden, J.S. End-on imaging: a new perspective on dorsoventral development in Drosophila embryos. Dev. Dyn. 237, 3252–3259 (2008).
Markow, T.A., Beall, S. & Matzkin, L.M. Egg size, embryonic development time and ovoviviparity in Drosophila species. J. Evolution. Biol. 22, 430–434 (2009).
Aboobaker, A.A., Tomancak, P., Patel, N., Rubin, G.M. & Lai, E.C. Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc. Natl Acad. Sci. USA 102, 18017–18022 (2005).
Ashburner, M. Grape-apple juice agar plates. In Drosophila Protocols (eds. Ashburner, M., Hawley, R.S. & Sullivan, W.) 658 (Cold Spring Harbor Laboratory Press, 2000).
Acknowledgements
This work was supported by the US National Science Foundation (DBI-0649833 to H.L., EFRI 1136913 to S.Y.S. and H.L.) and the US National Institutes of Health (grant nos. NS058465 to H.L. and GM078079 to S.Y.S.). H.L. is a DuPont Young Professor and a Sloan Research Fellow. We thank J. Ding for technical assistance.
Author information
Authors and Affiliations
Contributions
T.J.L. and M.Z. fabricated the devices and tested the protocol. H.L. and S.Y.S. oversaw the project. T.J.L. wrote the paper. All authors (T.J.L., M.Z.,B.L., S.Y.S. and H.L.) contributed to the design of the experiments and edited the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Microfluidic device layout and specifications (PDF 465 kb)
Rights and permissions
About this article
Cite this article
Levario, T., Zhan, M., Lim, B. et al. Microfluidic trap array for massively parallel imaging of Drosophila embryos. Nat Protoc 8, 721–736 (2013). https://doi.org/10.1038/nprot.2013.034
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2013.034
This article is cited by
-
Microfluidics for understanding model organisms
Nature Communications (2022)
-
A robot-assisted acoustofluidic end effector
Nature Communications (2022)
-
An open-source semi-automated robotics pipeline for embryo immunohistochemistry
Scientific Reports (2021)
-
Design of a comprehensive microfluidic and microscopic toolbox for the ultra-wide spatio-temporal study of plant protoplasts development and physiology
Plant Methods (2019)
-
Tools to reverse-engineer multicellular systems: case studies using the fruit fly
Journal of Biological Engineering (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.