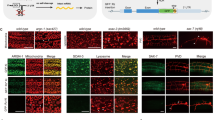
Abstract
We describe a method for conditional regulation of gene expression based on the processing of an intron cassette. The RNA processing factor MEC-8 is necessary for the function of the Caenorhabditis elegans touch receptor neurons; mec-8 mutants are touch insensitive. We show here that this insensitivity involves the loss of MEC-8–dependent splicing of mec-2, which encodes a component of the mechanosensory transduction complex. MEC-8 is needed to remove the ninth intron in mec-2 pre-mRNA to form the longest of three mRNAs, mec-2a. Without MEC-8, splicing causes the termination of the transcript. Inclusion of mec-2 intron 9 is sufficient to convey mec-8–dependent regulation on other genes and, in mec-8(u218ts) mutants, resulted in their temperature-dependent expression. Because mec-8 is expressed ubiquitously in embryos and extensively in larvae, this system should produce temperature-sensitive expression for most genes. As an example, we report a strain that exhibits temperature-dependent RNA interference.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Sauer, B. Inducible gene targeting in mice using the Cre/lox system. Methods 14, 381–392 (1998).
Dohmen, R.J., Wu, P. & Varshavsky, A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263, 1273–1276 (1994).
Gossen, M. & Bujard, H. Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. USA 89, 5547–5551 (1992).
Bacaj, T. & Shaham, S. Temporal control of cell-specific transgene expression in Caenorhabditis elegans. Genetics 176, 2651–2655 (2007).
Davis, M.W., Morton, J.J., Carrol, D. & Jorgensen, E.M. Gene activation using FLP recombinase in C. elegans. PLoS Genet. 4, e1000028 (2008).
Voutev, R. & Hubbard, A.J. A “FLP-out” system for controlled gene expression in Caenorhabditis elegans. Genetics 180, 103–119 (2008).
Horowitz, N.H. & Leupold, U. Some recent studies bearing on the one gene one enzyme hypothesis. Cold Spring Harb. Symp. Quant. Biol. 16, 65–74 (1951).
Epstein, R.H. et al. Physiological studies of conditional lethal mutants of bacteriophage T4D. Cold Spring Harb. Symp. Quant. Biol. 28, 375–394 (1963).
Chalfie, M. & Au, M. Genetic control of differentiation of the Caenorhabditis elegans touch receptor neurons. Science 243, 1027–1033 (1989).
Chalfie, M. & Sulston, J. Developmental genetics of the mechanosensory neurons of Caenorhabditis elegans. Dev. Biol. 82, 358–370 (1981).
Lundquist, E.A. et al. The mec-8 gene of C. elegans encodes a protein with two RNA recognition motifs and regulates alternative splicing of unc-52 transcripts. Development 122, 1601–1610 (1996).
Huang, M., Gu, G., Ferguson, E.L. & Chalfie, M. A stomatin-like protein necessary for mechanosensation in C. elegans. Nature 378, 292–295 (1995).
O'Hagan, R., Chalfie, M. & Goodman, M.B. The MEC-4 DEG/ENaC channel of Caenorhabditis elegans touch receptor neurons transduces mechanical signals. Nat. Neurosci. 8, 43–50 (2005).
Spike, C.A., Davies, A.G., Shaw, J.E. & Herman, R.K. MEC-8 regulates alternative splicing of unc-52 transcripts in C. elegans hypodermal cells. Development 129, 4999–5008 (2002).
Tavernarakis, N., Driscoll, M. & Kyrpides, N.C. The SPFH domain: implicated in regulating targeted protein turnover in stomatins and other membrane-associated proteins. Trends Biochem. Sci. 24, 425–427 (1999).
Davies, A.G., Spike, C.A., Shaw, J.E. & Herman, R.K. Functional overlap between the mec-8 gene and five sym genes in Caenorhabditis elegans. Genetics 153, 117–134 (1999).
Tabara, H. et al. The rde-1 gene, RNA interference, and transposon silencing in C. elegans. Cell 99, 123–132 (1999).
Sijen, T., Steiner, F.A., Thijssen, K.L. & Plasterk, R.H. Secondary siRNAs result from unprimed RNA synthesis and form a distinct class. Science 315, 244–247 (2007).
Goodman, M.B. et al. MEC-2 regulates C. elegans DEG/ENaC channels needed for mechanosensation. Nature 415, 1039–1042 (2002).
Lisbin, M.J., Qiu, J. & White, K. The neuron-specific RNA-binding protein ELAV regulates neuroglian alternative splicing in neurons and binds directly to its pre-mRNA. Genes Dev. 15, 2546–2561 (2001).
Zhou, H.L., Baraniak, A.P. & Lou, H. Role for Fox-1/Fox-2 in mediating the neuronal pathway of calcitonin/calcitonin gene-related peptide alternative RNA processing. Mol. Cell. Biol. 27, 830–841 (2007).
Poon, V.Y., Klassen, M.P. & Shen, K. UNC-6/netrin and its receptor UNC-5 locally exclude presynaptic components from dendrites. Nature 455, 669–673 (2008).
Zhang, S., Ma, C. & Chalfie, M. Combinatorial marking of cells and organelles with reconstituted fluorescent proteins. Cell 119, 137–144 (2004).
Chelur, D.S. & Chalfie, M. Targeted cell killing by reconstituted caspases. Proc. Natl. Acad. Sci. USA 104, 2283–2288 (2007).
Poyurovsky, M.V. et al. Nucleotide binding by the Mdm2 RING domain facilitates Arf-independent Mdm2 nucleolar localization. Mol. Cell 12, 875–887 (2003).
Yoshimatsu, T. & Nagawa, F. Control of gene expression by artificial introns in Saccharomyces cerevisiae. Science 244, 1346–1348 (1989).
Fu, G. et al. Female-specific insect lethality engineered using alternative splicing. Nat. Biotechnol. 25, 353–357 (2007).
Samson, M.L., Lisbin, M.J. & White, K. Two distinct temperature-sensitive alleles at the elav locus of Drosophila are suppressed nonsense mutations of the same tryptophan codon. Genetics 141, 1101–1111 (1995).
Soller, M. & White, K. ELAV inhibits 3′-end processing to promote neural splicing of ewg pre-mRNA. Genes Dev. 17, 2526–2538 (2003).
Jensen, K.B. et al. The tetranucleotide UCAY directs the specific recognition of RNA by the Nova K-homology 3 domain. Proc. Natl. Acad. Sci. USA 97, 5740–5745 (2000).
Charlet-B., N., Logan, P., Singh, G. & Cooper, T.A. Dynamic antagonism between ETR-3 and PTB regulates cell type-specific alternative splicing. Mol. Cell 9, 649–658 (2002).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Driscoll, M. & Chalfie, M. The mec-4 gene is a member of a family of Caenorhabditis elegans genes that can mutate to induce neuronal degeneration. Nature 349, 588–593 (1991).
Mello, C.C., Kramer, J.M., Stinchcomb, D. & Ambros, V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 10, 3959–3970 (1991).
Okkema, P.G. & Fire, A. The Caenorhabditis elegans NK-2 class homeoprotein CEH-22 is involved in combinatorial activation of gene expression in pharyngeal muscle. Development 120, 2175–2186 (1994).
Finney, M. & Ruvkun, G. The unc-86 gene product couples cell lineage and cell identity in C. elegans. Cell 63, 895–905 (1990).
Zhang, S. et al. MEC-2 is recruited to the putative mechanosensory complex in C. elegans touch receptor neurons through its stomatin-like domain. Curr. Biol. 14, 1888–1896 (2004).
Timmons, L. & Fire, A. Specific interference by ingested dsRNA. Nature 395, 854 (1998).
Acknowledgements
We thank M. Huang for providing valuable insight in the work with mec-2, and L. Chasin, J. Manley and members of the Chalfie laboratory for helpful discussions and comments on the manuscript. This work was supported by US National Institutes of Health grant GM30997 to M.C.
Author information
Authors and Affiliations
Contributions
A.C., C.M. and M.C. designed experiments; A.C. performed the experiments; A.C. and M.C. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Text and Figures
Supplementary Table 1 (PDF 72 kb)
Rights and permissions
About this article
Cite this article
Calixto, A., Ma, C. & Chalfie, M. Conditional gene expression and RNAi using MEC-8–dependent splicing in C. elegans. Nat Methods 7, 407–411 (2010). https://doi.org/10.1038/nmeth.1445
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1445
This article is cited by
-
Role of the ubiquitin–proteasome system in nervous system function and disease: using C. elegans as a dissecting tool
Cellular and Molecular Life Sciences (2012)
-
Knock it down, switch it on
Nature Methods (2010)