Abstract
The clinical significance of partial chromosome 3 alteration in uveal melanoma is still not clear. Also, the reported frequencies vary considerably in the published literature from 0 to 48%. The aims of the following study were to identify the frequency, molecular pathology and potential clinical significance of partial chromosome 3 alteration in uveal melanoma. We studied 47 uveal melanomas with an average follow-up of 36 months. Of these, 14 had confirmed metastasis. Allelic imbalance/loss of heterozygosity was studied using microsatellite markers on chromosome 3 enriched in markers located in the previously reported smallest regions of deletion overlap. Chromosomal alterations were assessed by conventional cytogenetics or comparative genomic hybridization (CGH) in a subset of patients. Utilizing genotyping, partial chromosome 3 alteration was detected in 14/47 tumors (30%). In the 23 tumors with available cytogenetic/CGH, partial chromosome 3 alteration was detected in 8/23 (38%) and was caused by both gains (4/8) and losses (4/8) of chromosome 3 with high frequency of complex chromosome 3 aberrations detected by cytogenetics. Out of the 14 tumors with confirmed metastasis, only 1 showed partial chromosome 3 alteration and the remaining showed monosomy 3. By limiting the aggressive disease marker to monosomy 3, genotyping showed 93% sensitivity and 67% specificity for detection of aggressive uveal melanoma. In conclusion, partial chromosome 3 alterations are common in uveal melanoma and mostly caused by complex cytogenetic changes leading to partial gains and/or partial losses of chromosome 3. Partial chromosome 3 alteration is not likely to be associated with highly aggressive uveal melanoma that metastasizes within the first 3 years after treatment. Microsatellite-based genotyping of chromosome 3 is highly sensitive for detection of aggressive uveal melanoma.
Similar content being viewed by others
Main
Monosomy of chromosome 3 is one of the most common genetic alterations in uveal melanoma with reported frequencies ranging between 30 and 50%.1, 2 The strong association between monosomy 3 and short survival, first reported by Prescher et al,3 has been well confirmed.4, 5, 6, 7 These studies have also shown that metastasis in the absence of monosomy 3 is quite rare. Based on such findings, monosomy 3 is currently being used in many clinical centers as a prognostic marker to identify aggressive uveal melanomas.1, 2, 8
Contrary to the well-documented clinical significance of monosomy 3, little is known about the clinical significance of partial chromosome 3 alterations in uveal melanoma. Two different approaches have been followed for analysis of partial chromosome 3 alterations in published literature. Some investigators excluded these lesions from their analysis based on the likelihood that they represent a different clinical subgroup,4, 9 whereas others grouped them with monosomy 3.10
The frequency of partial chromosome 3 alteration in uveal melanoma varies markedly in the published reports from 0 to 48% (Table 1).4, 9, 11, 12, 13, 14, 15, 16, 17, 18, 19 Although the vast majority of authors considered that it is caused by small deletions,4, 15, 17, 18, 20 it should be noted that genotyping, which was utilized by most of these reports, cannot reliably differentiate between deletions and other molecular mechanisms causing allelic imbalances, including mitotic recombination, aneuploidy and amplification.21 Few reports showed that partial chromosome 3 gains14 and unbalanced translocations15 could be involved in the pathogenesis of partial chromosome 3 alteration in uveal melanoma.
The aims of this study were to identify the frequency, clinical significance and potential molecular pathogenesis of partial chromosome 3 alteration in uveal melanoma.
Materials and methods
Samples
Primary uveal melanoma samples were collected from patients who underwent enucleation as the only treatment for their primary tumors at the Department of Ophthalmology, Ohio State University. A total of 47 tumors were included. The study was carried out in accordance with the protocol approved by the institutional review board . Table 2 summarizes the clinical data of the patients included in the study.
DNA Extraction and Genotyping
Tumor DNA was extracted from fresh frozen tumor tissue (39 samples) or from archival material (8 samples). Non-tumor DNA was extracted from either the peripheral blood lymphocytes (34 samples) or from nearby nontumor tissue (13 samples) using DNeasy tissue kit (Qiagen, Valencia, CA, USA). The DNA integrity was assessed using minigel electrophoresis. Extracted DNA was stored at 4 °C until the time of experiments. Genotyping was carried out using 16 microsatellite markers on chromosome 3, with 8 on each chromosome arm. Also, 12 additional markers were utilized in samples with partial chromosome 3 alterations. Microsatellite markers utilized were enriched in markers located within the smallest regions of overlap on chromosome 3 reported by other investigators.15, 17, 18 All genotyping markers were fluorescent labeled and either obtained from a commercial supplier (Research Genetics, Carlsbad, CA, USA) or custom made (SIGMA, St Louis, MO, USA) utilizing reported primer sequences (http://genome.ucsc.edu). Custom-made primers were labeled at the 5′ end with either FAM or HEX fluorescent dyes.
PCR conditions for all of these markers were heat activation for 15 min at 95 °C followed by 45 cycles of 94 °C for 30 s, 53 °C for 45 s, 72 °C for 1 min and final elongation at 72 °C for 10 min.
Genotyping was carried out at the Nucleic Acids Shared Resource, Comprehensive Cancer Center, Ohio State University. Briefly, fluorescently labeled PCR products were loaded in 7 μl hi-di formamide containing 1.2% LIZ500 size standard and were run on 3730 DNA analyzers. The data were analyzed using GeneMapper v3.5 software (Applied Biosystems, Foster City, CA, USA).
The allelic imbalance factor was determined by calculating the ratio of allele heights for both the normal (N) and tumor (T) sample, and then the tumor ratio was divided by the normal ratio=(T1:T2/N1:N2) as previously suggested.22, 23 To obtain positive ratios, T1 was selected to be the highest of the two alleles. Loss of heterozygosity was defined as total loss of one allele at a constitutional heterozygous locus22 or an allelic imbalance factor of ≥5. This ratio represents total loss of an allele observed in at least 80% of the tumor cells. Allelic imbalance was defined as a skewed intensity ratio between two alleles at a locus in the tumor compared with the normal DNA.22 We selected a threshold of allelic imbalance factor of >1.5 for scoring regions with allelic imbalance. This ratio is equivalent to loss of an allele observed in at least 33% of the tumor cells.
Conventional Cytogenetics
Karyotyping was carried out at the Cancer Cytogenetics Laboratory, Department of Pathology, Ohio State University according to routine protocols. Karyotyping was carried out for all prospectively collected tumors with sufficient amount of tumor tissue, a total of 20 tumors. Briefly, sterile collected tumor samples were finely minced and treated with collagenase for 1–4 h, until the cells were dissociated. The dissociated cells were cultured in RPMI-1640 (Gibco Invitrogen, Grand Island, NY, USA) +10% fetal bovine serum (Hyclone Laboratories, Logan, UT, USA), supplemented with L-glutamine, penicillin, streptomycin and insulin-transferrin-selenium (Gibco Invitrogen) and in MEM (Gibco Invitrogen) +10% fetal bovine serum supplemented with penicillin, streptomycin, L-glutamine and bone marrow condiment (BMC; Fisher Scientific, Houston, TX, USA). They were cultured on coverslips and in flasks until partially confluent. Harvest, GTW-banding and analysis were by standard laboratory procedures.
Comparative Genomic Hybridization (CGH)
CGH analysis was performed on 12 retrospectively collected tumors with available frozen tissue according to a published protocol24 using a CGH nick translation labeling kit (Vysis, Downers Grove, IL, USA). Normal reference DNA was labeled with Texas Red-5-dUTP (Applied Biosystems) and tumor DNA with Fluorescein-12-dUTP (Applied Biosystems). Hybridization of normal female DNA against normal male DNA was used as a negative control. CGH images were acquired with a cooled CCD camera attached to a Zeiss Axioplan 2 epifluorescence microscope, and analyzed using ISIS software (version 1.5.5) from Meta-Systems. We used fixed thresholds of >1.15 for gains and <0.85 for losses based on a two-color control male-to-female hybridization experiment. Data from at least 10 representative images of each chromosome were combined to generate an average ratio profile reflecting the relative copy number changes in test DNA.24
Statistical Analysis
Statistical analysis was carried out using STATA version 10 and GB-STAT software. Fisher's exact test was conducted to compare predictors of metastasis across molecular genetic prognostic markers, as well as other clinical prognostic factors such as gender, cell type, tumor size and extrascleral extension. Diagnostic accuracy calculations (sensitivity, specificity, positive and negative predictive values and likelihood ratios) were carried out on patients followed up for at least 36 months or with confirmed metastasis (total 32 participants). Kaplan–Meier curves were plotted to compare the median survival times for metastasis across the different categories of the independent variables (tumor molecular genetics and clinical prognostic criteria). Cox hazard regression was performed to calculate the hazard ratio associated with the previously mentioned variables.
Results
Frequency of Partial Chromosome 3 Alteration in Uveal Melanoma
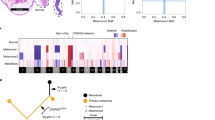
Genotyping identified monosomy 3 with all informative markers showing allelic imbalance/loss of heterozygosity in 26/47 (55%) of the samples included in the study. Allelic imbalances involving only parts of chromosome 3 with the remaining parts showing retention of heterozygosity was identified in 14 (30%) tumors (Figure 1). Heterodisomy of chromosome 3 with all the markers showing retention of heterozygosity was detected in 7 (15%) tumors.
Summary of genotyping of cases with partial chromosome 3 alterations. (a) Summary of genotyping: black boxes represent markers with allelic imbalance/loss of heterozygosity, white boxes represent markers with retention of heterozygosity; Ni: noninformative. (b) Chromosome 3 karyotypes of the four cases with partial chromosome 3 alteration and available cytogenetics (genotyping are marked by asterisks in a). (c) Representative case (UM9001), showing retention of heterozygosity in markers in p arm, allelic imbalance in D3S3045 marker on the 3q12 region and loss of heterozygosity in two other markers on q arm. P3: Partial chromosome 3 (deletion); P3*: partial chromosome 3 (gain).
Similar frequencies were observed in the subset of samples with available cytogenetics and CGH. Of the 11 samples with available cytogenetics, monosomy 3 was detected in 6 samples (55%), partial chromosome 3 alteration in 3 samples (27%) and heterodisomy 3 in 2 samples (18%) (Table 3). Of the 12 samples with available CGH, 7 samples (58%) showed monosomy 3, 4 samples showed partial chromosome 3 alterations (33%) and 1 sample showed disomy of chromosome 3 (8%).
Cytogenetics and CGH confirmed partial chromosome 3 alterations observed by genotyping in all but one of the 23 samples studied (Tables 3 and 4). In that sample, UM8004, genotyping identified partial chromosome 3 alteration in the form of allelic imbalance in markers from 3q13 to 3q27, whereas the rest of markers showed retention of heterozygosity. Cytogenetics revealed two normal chromosomes 3, with a marker chromosome suggesting that the marker chromosome likely contains part of chromosome 3.
Molecular Pathology of Partial Chromosome 3 Alteration in Uveal Melanoma
Chromosomal status, either by cytogenetics (ie, karyotyping) or CGH, was available in 8 of 14 samples with partial allelic imbalance/loss of heterozygosity. In four of these eight cases, partial losses were the cause, whereas in the remaining four cases gains of additional material were the cause of allelic imbalances of chromosome 3. Cytogenetics revealed chromosomal rearrangement involving chromosome 3 in three out of the four samples with partial chromosome 3 alterations. In the remaining sample, UM8004, no alteration of chromosome 3 was identified but a marker chromosome was detected (Table 3).
The majority, 12/14, of tumors with partial chromosome 3 alterations showed an allelic imbalance factor of <1.7, indicating genetic alteration in <41% of the tumor cells. Only three tumors with partial chromosome 3 alterations showed areas with allelic imbalance factor of ≥5. This was contrary to tumors with monosomy 3 where the majority 21/26 showed either total loss of an allele or an allelic imbalance factor of ≥5, indicating that the genetic alteration existed in >80% of the tumor cells. The difference was statistically significant (Fisher's exact test, P<0.0001). This observation suggests that in the majority of tumors with partial chromosome 3 alteration, the frequency of tumor heterogeneity, including tumor cells lacking genetic alteration in chromosome 3, is much higher than in monosomy 3 tumors.
Clinical Significance of Partial Chromosome 3 Alterations
The clinical significance of partial chromosome 3 alteration was assessed in a subset of patients with confirmed metastasis (14 patients) or a clinical follow-up of at least 36 months (additional 18 patients), using metastasis as the marker for aggressive disease. Of the 14 patients with confirmed metastasis, only one patient showed partial chromosome 3 alteration with loss of chromosome material on both the p and q arms (Figure 2). The difference in the incidence of metastasis between patients with monosomy 3 (13/19) and partial chromosome 3 alteration (1/9) was statistically significant (P=0.005, Fisher's exact test). Also, Kaplan–Meier analysis showed a significant association of monosomy 3, but not partial chromosome 3 alteration, with aggressive disease (log rank test, P=0.01, Figure 3). Of the eight tumors with partial chromosome 3 alteration and >36 months of metastasis-free survival (mean 67 months, range 39–119 months), 3 showed partial gains and 5 had partial losses. Tumors from four of the patients with partial loss showed allelic imbalance in, or nearby, the smallest region of deletion overlap previously reported in uveal melanoma.
Pathology and molecular genetics of the partial chromosome 3 case (UM4033) with confirmed metastasis. (a) A young patient (27 years old) with a bi-lobed tumor with different histological features. (b) Representative of spindle parts of the tumor. (c) Representative of epitheliod parts of the tumor. (d) CGH of chromosome 3 showing deletions (red bars) involving part of the p arm and the whole q arm. (e) Genotyping of markers from different areas of the tumor, tumor area (1) showing total allelic loss in both markers, whereas tumor area (2) showing allelic imbalance in the D3S4545 marker with total allelic loss in the D3S2418 marker. This indicates tumor heterogeneity with part of the tumor showing monosomy 3 and other part showing partial chromosome 3 alteration.
We also assessed the sensitivity and specificity of genotyping for detection of aggressive disease (Table 5). Combining monosomy 3 and partial chromosome 3 alterations as markers of tumor aggressiveness yielded 100% sensitivity but only 28% specificity for prediction of patients who developed metastasis. Limiting the assay to monosomy 3 with exclusion of partial chromosome 3 alterations slightly diminished the sensitivity to 93% but improved the assay specificity to 67%, whereas inclusion of samples with partial chromosome 3 alterations with areas of allelic imbalance factor of ≥5 (two tumors) showed 100% sensitivity and 61% specificity for detection of aggressive disease. These results suggest that the majority of partial chromosome 3 alterations are not associated with aggressive disease.
Other clinical prognostic factors such as tumor size, tumor location, extrascleral extension and cell type were not significantly associated with tumor aggressiveness (Table 6).
Discussion
The frequency of partial change in uveal melanoma varies considerably in the published literature (Table 1). Also, its clinical significance is still largely unknown. With the increasing utilization of molecular genetic testing for prediction of aggressive uveal melanoma, especially those based on the status of chromosome 3, understanding the frequency and clinical significance of partial chromosome 3 alterations is crucial for proper management of these patients. Our results indicate that the majority of partial chromosome 3 alterations, especially the cases due to gains of chromosomal materials, are not associated with aggressive disease. Only 1 of the 14 tumors with confirmed metastasis showed partial chromosome 3 alteration, whereas the remaining aggressive tumors showed allelic losses of all the markers on chromosome 3. The partial chromosome 3 case with metastatic disease was rather unique, with a bi-lobed tumor showing molecular genetic heterogeneity with monosomy 3 in one of the two tumor lobes whereas the other part of the tumor showed partial chromosome 3 loss (Figure 2).
There are few reports of metastatic uveal melanoma showing partial chromosome 3 alterations.4, 9, 15 Utilizing genotyping, Scholes et al4 reported one case who died of metastatic disease; however, no further information was presented as the authors excluded all partial chromosome 3 cases from their detailed analysis. Cross et al,15 also utilizing genotyping, reported one case with partial chromosome 3 involving the 3pter–3q12 region who died of metastatic disease. Conventional cytogenetics of that case showed a near-triploid karyotype with two apparently normal copies of chromosome 3 and several marker chromosomes,15 suggesting a chromosome 3 parental isodisomy with additional material on chromosome 3. In that study, two additional cases with partial chromosome 3 alterations were metastasis free for 60 months, whereas another two were metastasis free for 23 months.15 Trolet et al,9 using array CGH, reported four cases of partial chromosome 3 losses who developed metastatic disease and several metastatic uveal melanoma lesions with partial chromosome 3 losses. The apparent discrepancy between the lack of association between partial chromosome 3 alterations and metastatic disease in our study and that of Trolet et al9 could be because of the relative smaller sample size in our study or to our relative short follow-up duration. As the vast majority of metastases from uveal melanomas occur during the first 3 years after treatment of the primary tumor,25 studies utilizing 3-year follow-up period, similar to ours, should identify highly aggressive, rapidly metastasizing tumors. It is worth noting that the original publications linking monosomy 3 to aggressive uveal melanoma utilized 3-year follow-up duration.3, 26 Also, studies linking gene expression-based molecular classifiers to tumor aggressiveness utilized similar follow-up duration.12 The unusual high frequency of metastasizing disomy 3 tumors reported by Trolet et al9 could be related to the follow-up duration that was not specified in their report.9 It is worth noting that we identified partial chromosome 3 alteration in the metastatic lesion of a patient who developed metastatic disease 20 years after treatment of the primary tumor (data not shown). Whether the molecular genetics of rapidly metastatic uveal melanoma is different from the slowly metastatic disease still remains to be investigated.
Our study indicates that the frequency of partial chromosome 3 alteration is rather high in uveal melanoma. However, such frequency varies widely in the published literature and ranges from 0 to 48% (Table 1). Potential explanations of such variation include the methodology utilized, threshold for detecting genetic alteration, size of the tissue fragment studied, sample fixation, difference in ethnicity, tumor size, tumor heterogeneity or a combination of any of these factors. For example, the marked variation of the frequency of partial chromosome 3 alteration between the original report of Parrella et al19 (8.5%) and their later report17 (34.7%) utilizing the same samples was because of the inclusion of a considerably larger number of genotyping markers in their later study. Similarly, the utilization of different techniques and different thresholds for assessment of the genetic alteration could explain the large difference between the 5% of partial chromosome 3 alteration reported by Scholes et al4 in their report utilizing genotyping, and the high frequency ranging from 28.8 to 48% reported in their later study on the same samples utilizing multiplex ligation-dependent probe amplification.11
Tumor heterogeneity27, 28, 29 and the sensitivity of the molecular technique to identify such heterogeneity, including the threshold utilized for detection of genomic alteration, are additional factors explaining variation in the frequency of partial chromosome 3 alteration between different studies. It is worth noting that in the majority (80.7%) of our monosomy 3 cases, we observed either total loss of an allele or an allelic imbalance factor of ≥5, indicating that the genetic alteration existed in >80% of the tumor cells. This could be because of either deletion of one copy of chromosome 3 or loss and reduplication leading to acquired parental isodisomy. Such genetic change could be easily detected using various molecular techniques. On the other hand, the majority of cases with partial chromosome 3 alteration (85.7%) showed low allelic imbalance factor (<1.7), indicating significant heterogeneity of the tumors. Molecular techniques and sampling methodologies that include tumor DNA obtained from a relatively large portion of tumor tissue, such as whole tissue sections, are more capable of detecting tumor heterogeneity compared with methods utilizing only small portion of tumor tissues, such as needle biopsy or small fragments from snap-frozen tumor tissues.
The molecular pathogenesis of partial chromosome 3 alterations in uveal melanoma is largely believed to be caused by small deletions.4, 15, 17, 18, 20 Based on that, several smallest regions of deletion overlap have been reported.15, 17, 18 These areas include 3p24–26, 3p11–14,3q (Cross et al15), 3p25.1–3p25.2 (Parrella et al17), 3p25 and 3q24–q26 (Tschentscher et al18). It has been suggested that such smallest regions of deletion overlap are the location of tumor-suppressor genes important in the pathogenesis of metastasizing uveal melanoma. It should be noted that of the three reports defining these regions, only two utilized a combination of copy number alteration and allelic imbalances,15, 18 whereas the third utilized only genotyping.17 Samples with small gains could bias the assessment as genotyping cannot differentiate between areas of gains and losses at low allelic imbalance factor thresholds. In fact, several of our cases with confirmed partial gains showed allelic imbalances in the reported smallest region of deletion overlap by genotyping.
Our study indicates the high sensitivity of microsatellite-based genotyping for detection of highly aggressive uveal melanoma. Similar results were reported by other investigators utilizing single-nucleotide polymorphism.13 Microsatellite genotyping is readily available in the majority of clinical molecular laboratories and can be adapted for clinical testing. It is worth noting that our study, similar to the vast majority of other reports on partial chromosome 3, utilized only uveal melanoma treated by enucleation. Further validation of our findings including studies of molecular genetic changes in biopsied small- and medium-sized tumors is recommended.
In conclusion, our results suggest that partial chromosome 3 alteration is common in uveal melanoma. The molecular mechanisms for such changes include complex chromosomal re-arrangements, partial gains and partial deletions. The majority of partial chromosome 3 alterations are not associated with metastatic disease, in particular, rapidly aggressive tumors.
References
Damato B, Coupland SE . Translating uveal melanoma cytogenetics into clinical care. Arch Ophthalmol 2009;127:423–429.
Shields JA, Shields CL, Materin M, et al. Role of cytogenetics in management of uveal melanoma. Arch Ophthalmol 2008;126:416–419.
Prescher G, Bornfeld N, Hirche H, et al. Prognostic implications of monosomy 3 in uveal melanoma. Lancet 1996;347:1222–1225.
Scholes AG, Damato BE, Nunn J, et al. Monosomy 3 in uveal melanoma: correlation with clinical and histologic predictors of survival. Invest Ophthalmol Vis Sci 2003;44:1008–1011.
Patel KA, Edmondson ND, Talbot F, et al. Prediction of prognosis in patients with uveal melanoma using fluorescence in situ hybridisation. Br J Ophthalmol 2001;85:1440–1444.
Aalto Y, Eriksson L, Seregard S, et al. Concomitant loss of chromosome 3 and whole arm losses and gains of chromosome 1, 6, or 8 in metastasizing primary uveal melanoma. Invest Ophthalmol Vis Sci 2001;42:313–317.
White VA, Chambers JD, Courtright PD, et al. Correlation of cytogenetic abnormalities with the outcome of patients with uveal melanoma. Cancer 1998;83:354–359.
Harbour JW . Molecular prognostic testing and individualized patient care in uveal melanoma. Am J Ophthalmol 2009;148:823–829 e821.
Trolet J, Hupe P, Huon I, et al. Genomic profiling and identification of high-risk uveal melanoma by array CGH analysis of primary tumors and liver metastases. Invest Ophthalmol Vis Sci 2009;50:2572–2580.
Shields CL, Ganguly A, Materin MA, et al. Chromosome 3 analysis of uveal melanoma using fine-needle aspiration biopsy at the time of plaque radiotherapy in 140 consecutive cases. Trans Am Ophthalmol Soc 2007;105:43–52; discussion 52–43.
Damato B, Dopierala J, Klaasen A, et al. Multiplex ligation-dependent probe amplification of uveal melanoma: correlation with metastatic death. Invest Ophthalmol Vis Sci 2009;50:3048–3055.
Worley LA, Onken MD, Person E, et al. Transcriptomic versus chromosomal prognostic markers and clinical outcome in uveal melanoma. Clin Cancer Res 2007;13:1466–1471.
Onken MD, Worley LA, Person E, et al. Loss of heterozygosity of chromosome 3 detected with single nucleotide polymorphisms is superior to monosomy 3 for predicting metastasis in uveal melanoma. Clin Cancer Res 2007;13:2923–2927.
White JS, McLean IW, Becker RL, et al. Correlation of comparative genomic hybridization results of 100 archival uveal melanomas with patient survival. Cancer Genet Cytogenet 2006;170:29–39.
Cross NA, Ganesh A, Parpia M, et al. Multiple locations on chromosome 3 are the targets of specific deletions in uveal melanoma. Eye 2006;20:476–481.
Hughes S, Damato BE, Giddings I, et al. Microarray comparative genomic hybridisation analysis of intraocular uveal melanomas identifies distinctive imbalances associated with loss of chromosome 3. Br J Cancer 2005;93:1191–1196.
Parrella P, Fazio VM, Gallo AP, et al. Fine mapping of chromosome 3 in uveal melanoma: identification of a minimal region of deletion on chromosomal arm 3p25.1-p25.2. Cancer Res 2003;63:8507–8510.
Tschentscher F, Prescher G, Horsman DE, et al. Partial deletions of the long and short arm of chromosome 3 point to two tumor suppressor genes in uveal melanoma. Cancer Res 2001;61:3439–3442.
Parrella P, Sidransky D, Merbs SL . Allelotype of posterior uveal melanoma: implications for a bifurcated tumor progression pathway. Cancer Res 1999;59:3032–3037.
Shields CL, Ganguly A, Materin MA, et al. Chromosome 3 analysis of uveal melanoma using fine-needle aspiration biopsy at the time of plaque radiotherapy in 140 consecutive cases. Arch Ophthalmol 2007;125:1017–1024.
Devilee P, Cleton-Jansen AM, Cornelisse CJ . Ever since Knudson. Trends Genet 2001;17:569–573.
Skotheim RI, Diep CB, Kraggerud SM, et al. Evaluation of loss of heterozygosity/allelic imbalance scoring in tumor DNA. Cancer Genet Cytogenet 2001;127:64–70.
Cawkwell L, Bell SM, Lewis FA, et al. Rapid detection of allele loss in colorectal tumours using microsatellites and fluorescent DNA technology. Br J Cancer 1993;67:1262–1267.
Kallioniemi A, Krahu R, Kallioniemi O-P . Comparative genomic hybridization. In: Taylor GR (ed). CRC Press: Boca Raton, New York, London, Tokyo, 1997, pp 273–285.
Singh AD, Topham A . Survival rates with uveal melanoma in the United States: 1973–1997. Ophthalmology 2003;110:962–965.
Sisley K, Rennie IG, Parsons MA, et al. Abnormalities of chromosomes 3 and 8 in posterior uveal melanoma correlate with prognosis. Genes Chromosomes Cancer 1997;19:22–28.
Mensink HW, Vaarwater J, Kilic E, et al. Chromosome 3 intratumor heterogeneity in uveal melanoma. Invest Ophthalmol Vis Sci 2009;50:500–504.
Maat W, Jordanova ES, van Zelderen-Bhola SL, et al. The heterogeneous distribution of monosomy 3 in uveal melanomas: implications for prognostication based on fine-needle aspiration biopsies. Arch Pathol Lab Med 2007;131:91–96.
Sandinha T, Farquharson M, McKay I, et al. Correlation of heterogeneity for chromosome 3 copy number with cell type in choroidal melanoma of mixed-cell type. Invest Ophthalmol Vis Sci 2006;47:5177–5180.
Acknowledgements
This work is funded by the Patti Blow Research fund in Ophthalmology and a Selective Investment grant from the College of Medicine, Ohio State University.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Rights and permissions
About this article
Cite this article
Abdel-Rahman, M., Christopher, B., Faramawi, M. et al. Frequency, molecular pathology and potential clinical significance of partial chromosome 3 aberrations in uveal melanoma. Mod Pathol 24, 954–962 (2011). https://doi.org/10.1038/modpathol.2011.51
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/modpathol.2011.51
Keywords
This article is cited by
-
Lack of BAP1 protein expression in uveal melanoma is associated with increased metastatic risk and has utility in routine prognostic testing
British Journal of Cancer (2014)
-
Exome sequencing identifies recurrent somatic mutations in EIF1AX and SF3B1 in uveal melanoma with disomy 3
Nature Genetics (2013)
-
Prognostic significance of chromosome 3 alterations determined by microsatellite analysis in uveal melanoma: a long-term follow-up study
British Journal of Cancer (2012)