Abstract
Integration of exogenous DNA into a host genome represents an important route to generate animal and cellular models for exploration into human disease and therapeutic development. In most models, little is known concerning structural integrity of the transgene, precise site of integration, or its impact on the host genome. We previously used whole-genome and targeted sequencing approaches to reconstruct transgene structure and integration sites in models of Huntington’s disease, revealing complex structural rearrangements that can result from transgenesis. Here, we demonstrate in the R6/2 mouse, a widely used Huntington’s disease model, that integration of a rearranged transgene with coincident deletion of 5,444 bp of host genome within the gene Gm12695 has striking molecular consequences. Gm12695, the function of which is unknown, is normally expressed at negligible levels in mouse brain, but transgene integration has resulted in cortical expression of a partial fragment (exons 8–11) 3’ to the transgene integration site in R6/2. This transcript shows significant expression among the extensive network of differentially expressed genes associated with this model, including synaptic transmission, cell signalling and transcription. These data illustrate the value of sequence-level resolution of transgene insertions and transcription analysis to inform phenotypic characterization of transgenic models utilized in therapeutic research.
Similar content being viewed by others
Introduction
Fundamental questions in human biology and therapeutic development have been evaluated using transgenic model organisms, typically generated by pronuclear injection. Despite its widespread use, relatively little is known concerning the detailed impact of random transgene integration on the host genome as a result of pronuclear injection, although there has been documented evidence of position effect variegation and insertional mutations disrupting endogenous genes1. Conventional methods to characterize transgene integrations (e.g. fluorescence in situ hybridization [FISH] or Southern blotting) lack the resolution to determine the integration site, and sequence-level integrity of both the transgene and host genome.
We have shown previously that customizations in next-generation sequencing methods can deliver unambiguous localization of transgenic integration sites and characterization of the transgene architecture at nucleotide level resolution. We applied these methods to a number of Huntington’s disease (HD) model organisms2, including the widely studied R6/2 transgenic mouse model3. This line was one of a series created in 1996 by pronuclear injection of a 1.9 kb fragment from the 5′ end of human HTT, containing the first exon with a ~130 unit CAG trinucleotide repeat expansion. In the R6/2 mouse, an amino-terminal polyglutamine-containing fragment of huntingtin is expressed and leads to rapid and severe neurological abnormalities at a younger age than other N-terminal HTT-fragment transgenic mouse models, making this mouse line attractive for therapeutic studies.
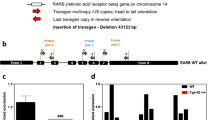
Using a series of targeted and whole-genome sequencing approaches, we previously revealed significant structural rearrangement of the transgene in R6/2 genomic DNA at nucleotide resolution, including excision/insertion and inversion events that left a single copy of the 5′ region and exon 1 CAG expansion region downstream of a 168 bp insertion of bacterial DNA and of intron 1 sequence, along with additional rearrangement of the upstream and intron 1 sequences (Fig. 1, see Chiang et al.2 for complete details, GenBank: KF990992.1). This structure suggests the R6/2 transgene integration event involved fragments derived from at least three copies of the original injected HTT fragment, as originally speculated by the authors who created the model3 and suggested by previous analyses4. However, we also found complex inversions, as well as head-to-tail and tail-to-head concatenations. This entire segment was inserted, with coincident deletion of 5,444 bp of host DNA, within intron 7 of Gm12695 in mouse chromosome 4. The transgene inserted in antisense orientation to Gm12695 transcription, deleting a segment predicted to harbour multiple transcription factor binding and DNaseI hypersensitivity sites that encompass two regions of greater than 90% identity across mammals (Fig. 1). Here, we further these studies by assessing the transcriptional impact of the transgenic integration in the R6/2 mouse.
(a) The original structure of the R6 transgene (top schematic, drawn to scale) and its extensive resultant rearrangement following integration into chromosome 4 of the R6/2 mouse (bottom schematic) are shown. The R6/2 integrated transgene contained fragments derived from at least three copies of the original R6 transgene, including complex inversions as well as head-to-tail and tail-to-head concatenations. Additionally, a 168 bp fragment of bacterial sequence was integrated upstream of the transgene HTT 5′ genomic sequence2. (b) Integration of a 1.9 kb HTT transgene fragment into intron 7 of mouse locus Gm12695 and concomitant 5.4 kb deletion of intron 7 at the integration site (demarcated by dotted lines). PhyloP38 analysis of mammalian conservation spanning the gene is provided, revealing the deleted intronic segment to be among the most conserved regions in the gene, including its coding sequence.
Results
To determine whether the transgene integration event in the R6/2 genome had an effect on expression of Gm12695 in cortical tissue, we initially used a quantitative RT-PCR approach. We found that Gm12695 is expressed at negligible levels in the cortex of non-transgenic (wild-type) littermates but showed evidence of dramatically increased levels, greater than 30-fold, in the corresponding brain tissues of R6/2 mice. There were at least two potential explanations for this increase; 1) the integration of the transgene within the noncoding intronic sequence of Gm12695 profoundly disrupts its normal regulation, resulting in altered expression of this gene in these regions of the brain, or 2) the increased expression of Gm12695 is an indirect consequence of the expression of the R6/2 transgene, as a reaction to the pathogenesis/toxicity caused by expression of the polyglutamine fragment. To distinguish between these options, we examined further mouse lines that are descended from the original R6/2 mouse line but show marked differences in their severity of phenotype due to substantial changes in the length of the CAG trinucleotide repeat. The repeat is known to be unstable in these mice3 both in somatic cells and in the germline, hence selective breeding has enabled an allelic series to be generated harbouring different CAG repeat lengths5, as determined by Laragen Inc.6. Mice with considerably longer repeats (404 CAGs, 714 CAGs) or shorter repeats (52 CAGs) display a less severe phenotype than the parental R6/2 strain (111 CAGs) from which they were generated, with a delay in symptom progression and prolonged survival5,7. We reasoned that if Gm12695 expression was a consequence of pathogenesis/toxicity induced by the expression of the polyglutamine fragment, its expression would vary across each model harbouring different CAG repeat lengths. If, on the other hand, its expression were independent of polyglutamine fragment pathogenesis/toxicity, we would expect its expression to remain unchanged across each of these models. We used the Pfaffl analysis method8 with primers spanning exons 8 and 9 of Gm12695 and normalized to the reference gene Atp5b. We found that Gm12695 is highly expressed in all four CAG repeat expansion mice (52, 111, 404 and 714 CAG repeats), in three different brain regions (cortex, hippocampus, striatum) (Fig. 2a,b,c), suggesting that the increased expression of Gm12695 is not a consequence of pathology/toxicity caused by expression of the transgene-encoded polyglutamine fragment. Expression analysis in two peripheral tissues, heart and liver, demonstrated the same aberrant expression of Gm12695 in R6/2 (Supplementary Fig. S1).
Expression of Gm12695 mRNA sequences in (a) cortex, (b) hippocampus and (c) striatum of four independent R6/2 transgenic mice expressing either 52, 111, 404 or 714 CAG repeats (which display differing phenotypic severity), and four age and sex-matched littermates that do not carry the transgene, was measured using quantitative PCR. All brain regions in transgenic R6/2 mice showed dramatically increased expression of Gm12695 sequences above low or negligible expression levels in wild-type. Data are represented as mean ± STDEV.
To explore the consequent alterations to normal transcription, both locally and globally of the transgene integration event, we performed RNA sequencing (RNAseq) on cortical brain tissue from three R6/2 mice (160 CAG+/−5) obtained from the Jackson Laboratory and three littermate controls. RNAseq revealed that the normal transcript structure of Gm12695 was significantly altered in R6/2 samples. Specifically, a portion of the 3′ end of the transgene (on the reverse strand) spliced to exons 8–11 of Gm12695 (sense strand) were joined, creating a possible open reading frame (see Fig. 3, Supplementary Fig. S2). This is consistent with the Q-PCR data, which targeted transcription from exons 8 and 9 of Gm12695. The transcriptome assembly tool, Trinity9, also predicted this transcript. The majority of transcript sequence reads were aligned to a region on the reverse strand of the transgene spanning chr4:96,743,050–96,743,250, which overlaps with bacterial DNA that was integrated into the transgene structure following pronuclear injection (Supplementary Table S1). Relatively few transcript reads aligned over the rest of the transgene on the reverse strand, including the CAG repeat region due to secondary structure of the RNA and the intractability of aligning long repetitive sequences (Supplementary Fig. S4).
Integrated Genome Viewer display of combined RNA sequencing data from all three R6/2 libraries. The Sashimi plot depicts splice junctions as arcs between the R6/2 transgene and exon 8, and between other exons of the Gm12695 transcript on the sense strand (reverse strand). Numbers on the arcs illustrate the reads covering splice junctions. Maximum read depth coverage is 89. The bottom track illustrates a novel transcript predicted by Trinity (predicted transcript) as a result of transgene integration, the known Gm12695 transcript and the R6/2 transgene at the integration site. The chromosomal coordinates are derived from Supplementary Table S1.
Minimal reads mapped to the forward strand of the transgene (antisense strand of Gm12695) (Supplementary Figs S3 and S5). However, downstream of the transgene insertion, in intron 7 of Gm12695 there was remarkably high levels of expression (Supplementary Fig. S3). This region is predicted to contain repeat elements including a long terminal repeat (LTR), MLT1A on the reverse strand and a long interspersed nuclear element (LINE), L1Md_T on the forward strand (Supplementary Fig. S5). Neither splice junctions nor mapped paired-end reads spanning both the transgene and repeat elements in Gm12695 were detected (Supplementary Figs S2 and S3), hence no new open reading frames were created. However, it is possible that transgene integration has activated expression of these elements, which are not normally transcribed in control mice. As predicted, we did not detect reads matching Gm12695 or the R6/2 transgene in the control mice on either strand.
To analyse whether expression of the transgene and partial expression of Gm12695 were associated with alteration of other gene networks in the R6/2 model, we used differential expression analysis to compare R6/2 and control samples. We detected 19,514 expressed genes among which 65 were significantly differentially expressed at adjusted p-value < 0.05 and 841 were differentially expressed at nominal p-value < 0.05 (Supplementary Tables S2 and S3). Pathway and gene ontology (GO) term overrepresentation analysis revealed that R6/2 down-regulated genes at nominal p < 0.05 were associated with terms related to neurobiology including “neurological system process”, “synaptic transmission”, “neuropeptide signalling pathway”, “synapse”, “voltage-gated ion channel activity”, “cell-cell signalling” and “calcium signalling pathway”, whereas R6/2 up-regulated genes at nominal p < 0.05 were enriched for terms associated with “transcription” (Supplementary Table S4). Not surprisingly, the partially transcribed Gm12695 (sense strand) was the most significant differentially expressed transcript (adjusted p-value = 6.66e-28), whereas differential expression of the transgene on the corresponding forward strand ranked 40th (adjusted p-value = 1.86e-2; Supplementary Table S3).
We also investigated Gm12695 expression using RNA sequencing data from heterozygous Htt CAG repeat expansion knock-in mice10, genetic replicas of the HD mutation. Members of this allelic knock-in series harbouring increasing size CAG repeat tracts (often named Q20, Q80, Q92, Q111 generated by Wheeler et al.11 and Q140, Q175 generated by Menalled et al.12,13) exhibit a variety of disease-allele associated phentoypes12,14,15,16,17. We found no evidence of Gm12695 expression across any of the expanded CAG repeat lengths, tissue types (both neuronal and non-neuronal) and ages for this allelic series (Supplementary Fig. S6). We also identified a recent HD post-mortem RNA sequencing dataset18 which explored global transcriptional dysregulation in HD and control prefrontal cortex. We found the human orthologue of this gene (C1orf87) to be upregulated in HD cortex, however this effect was removed when the dataset was adjusted for clinical covariates such as CAG repeat length and residual age of onset. These data both support the notion that aberrant expression of Gm12695 is specifically related to the transgene insertion event in R6/2 and is not an indirect consequence of HD pathology.
Discussion
Using Q-PCR and RNAseq we have shown the transcriptional impact of transgenic integration in a fragment model of Huntington’s disease, the R6/2 mouse model, one of the most widely studied models of the disease. Initial elucidation of the genomic insertion site, into the conserved gene Gm12695, revealed complex structural rearrangement, including a 5.4 kb deletion of host Gm12695 sequence. Q-PCR studies targeted downstream of the insertion site revealed this gene to be abnormally highly expressed in R6/2 brain across mice with varying phenotypic severity. This suggested that the increased expression of the host gene sequences was not an indirect reaction to the pathogenesis/toxicity caused by expression of the polyglutamine fragment, but rather due to the transgene integration event.
To clarify the impact of the transgene and concomitant deletion of the host sequence, we used RNAseq to reveal transcriptional activation of the downstream mouse Gm12695 exons 8–11, as well as expression of an upstream LINE sequence on the opposite strand. These changes in gene expression are associated with differentially expressed gene networks in R6/2 mice, including synaptic transmission, cell signalling and transcription. Thus, the integration of the transgene into intron 7 of Gm12695 appears to substantially disrupt its normal regulation, resulting in dysregulated expression of a conserved gene that is not normally expressed (or expressed at very low levels) in mouse brain. Transcriptional analysis of an HD mouse knock-in allelic series11,12,13 and a human post-mortem dataset18 did not reveal significant dysregulation of Gm12695 or its human orthologue C1orf87, supporting the finding that abnormal regulation of Gm12695 is a consequence of transgene integration in R6/2. Dysregulation of this gene has not been a focus of previous gene expression studies in this R6/2 mouse model. This may be because the gene is largely uncharacterized with unknown function, although the gene has been implicated in amyloid beta pathology in Alzheimer’s disease19,20.
Gm12695, like its conserved human orthologue, C1orf87, encodes an anonymous EF-hand protein of which little is known. It is not clear whether this abnormal expression of Gm12695 is due specifically to the insertion of the transgene sequence, the deletion of the 5.4 kb host Gm12695 sequence, or the combination of the two. This may be elucidated using a CRISPR-based approach to model the integration event in neuronal or induced pluripotent stem cells. We also cannot definitively disentangle whether this expression change contributes to any phenotypes shared by R6/2 and its sub-lines, or whether it acts as a modifier on the sensitized background of R6/2 to make the model more severe than other N-terminal fragment models.
These results suggest that it is advisable to determine the genomic integration site and structural architecture of introduced transgenes to understand fully the consequences of transgenesis. This can be readily accomplished by next-generation sequencing methods as we have reported with transgenic mouse and sheep models utilized in HD research2. Furthermore, the same methods can be applied to achieve the necessary high-resolution structural characterization for other genome integration phenomena, including viral insertions or the introduction of reprogramming transcription factors in induced pluripotent stem cell models. Capitalizing on the capability of current genomics technologies to characterize disease models in advance of costly functional studies may provide a valuable guide to choosing individual lines best suited for biological interpretation and therapeutic development.
Methods
Samples
R6/2 mouse mRNA was prepared from fresh frozen brain tissue (cortex, hippocampus and striatum) from either in-house colonies (R6/2 mice and their wild-type littermates) at the University of Cambridge or from tissue purchased from the Jackson Laboratory. The Jackson Laboratory is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC International). This study was carried out in accordance with the recommendations of the United Kingdom 1986 Animals (Scientific Procedures Act). The protocol was approved by the Ethical Review committee of the University of Cambridge. RNA was prepared for Quantitative PCR and RNA sequencing from ~30 mg of brain tissue, homogenized using QIAzol lysis reagent (Qiagen) in a Qiagen TissueLyser (2 × 2 min at 20 Hz). RNA was subsequently purified using Qiagen’s RNeasy Lipid Tissue Mini Kit following manufacturers instructions.
Quantitative PCR
Purified RNA was DNaseI treated with TURBO DNA-free Kit (Ambion) and quantified by NanoDrop. 400 ng RNA was used in the first-strand cDNA synthesis reaction primed with random hexamers using Invitrogen’s SuperScript III First-Strand Synthesis SuperMix. cDNA was diluted 1:10 in dH2O and 2.5 μl (striatum, hippocampus) or 1.5 μl (cortex) used per 10 μl real-time PCR reaction with 2x LightCycler 480 SYBR Green I Master and 0.5 μM of each primer. Primer sequences used for Gm12695 were: Gm12695 F 5′ cagagcttctgctgatttgtca 3′, Gm12695 R 5′ cagggatgtcaaccaagtcc 3′, and for the reference gene Atp5b21: Atp5b F 5′ tgagagaggtcctatcaaaacca 3′, Atp5b R 5′ caccagaatctcctgctcaac 3′. Reverse transcriptase negative controls were included for each sample and no-template controls performed for each primer pair. Samples were run in triplicate on a Roche LightCycler 480 2.0 Real-Time PCR System. After an initial denaturation at 95 °C for 5 minutes samples underwent 45 cycles of amplification at 95 °C for 15 seconds and 60 °C for 1 minute, with melting curve analysis performed at the end of each run. Standard curves were run for each primer pair in each tissue group and subsequently used in the relative quantification of Gm12695, using the Pfaffl method8. Gm12695 crossing point values for all wild-type samples were at least 5 cycles higher than transgenic samples (with all exceeding 32 cycles), and arbitrary values of 40 were given to those that did not express any Gm12695. Therefore, Gm12695 expression was quantified in transgenic animals by comparison with the combined wild-type mean.
RNA sequencing
Six RNA-sequencing libraries were prepared from cortical tissue extracted from three R6/2 transgenic female mice and three control mice (age, sex and strain matched from The Jackson laboratory) with a customized version of the strand-specific dUTP method22,23,24, as we have previously described25. One microliter of a 1:10 dilution of External RNA Controls Consortium (ERCC) spike-ins (Ambion) containing 92 synthetic RNA standards of known concentrations and sequence was added to each RNA-sequencing library to estimate the expression threshold to detect the expressed genes. Libraries were multiplexed, pooled and sequenced on Illumina HiSeq2000, generating an average of 34.4 million paired-end reads of 51 bp.
R6/2 Bioinformatic analysis
Quality of sequence reads was assessed by fastQC (v. 0.10.1)26. The transgene sequence of 2,463 nt. (GenBank:KF990992.1) was inserted into a mouse reference genome Ensembl GRCm38 (v. 75) by replacing the 5,444 bp-long segment spanning chr4:96,742,795–96,748,238. Sequence reads were then aligned to this modified mouse genome with an inserted transgene sequence using GSNAP (v. 12–19–2014) at its default parameter setting27. Quality checking of alignments was assessed by custom scripts utilizing Picard Tools (http://broadinstitute.github.io/picard/), RNASeQC28, RSeQC29 and SamTools30. Genome-guided transcriptome assembly for chromosome 4 was performed using Trinity (v 2.0.6) on combined alignments of all three R6/2 samples with parameters –SS_lib_type RF and –genome_guided_max_intron 1000031. Gene level counts were tabulated using BedTools’s multibamcov algorithm (v. 2.17.0)32 on unique alignments for each library at all Ensembl genes (GRCm38 v. 75) except Gm12695, for which a transcript predicted by Trinity was used. This transcript consisted of a portion of the transgene and exons 8–11 of Gm12695. Analysis of ERCC spike-ins as described in Blumenthal et al.33 estimated the expression threshold for detection to be at least two uniquely mapped reads. 19,514 genes that met this threshold in either all the control samples or all the R6/2 samples were further considered for differential expression analysis, which was carried out using nbinomTest function in an R/Bioconductor package, DESeq (v. 1.18) in R platform (v. 3.1.0). Over-representation analysis of the pathway and gene ontology (GO) terms was performed using DAVID (v. 6.7)34, where we used ensemble ids to represent genes and 19,514 expressed genes as a background gene set. Only terms with Benjamini-adjusted p value < 0.05 were reported. Alignments and splice junctions were visualized using the Integrative Genomics Viewer (v. 2.3.72)35.
Htt knock-in mice bioinformatic analysis
RNA sequence datasets for the Htt knock-in allelic series are available at NCBI GEO accession numbers: GSE73468, GSE73503, GSE73508, GSE65770, GSE65772, GSE65774, GSE65775, GSE65776. Trimmed fastq files were obtained and aligned to the mouse GRCm38.84 reference genome with STAR36 and quantitated at the gene level with HTSeq37. These counts were used to calculate FPKM and transcripts per million (TPM) thereby normalizing for both gene size and library size.
Additional Information
How to cite this article: Jacobsen, J. C. et al. Potential molecular consequences of transgene integration: The R6/2 mouse example. Sci. Rep. 7, 41120; doi: 10.1038/srep41120 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Woychik, R. P. & Alagramam, K. Insertional mutagenesis in transgenic mice generated by the pronuclear microinjection procedure. Int. J. Dev. Biol. 42, 1009–1017 (1998).
Chiang, C. et al. Complex reorganization and predominant non-homologous repair following chromosomal breakage in karyotypically balanced germline rearrangements and transgenic integration. Nat. Genet. 44, 390–397, doi: http://www.nature.com/ng/journal/v44/n4/abs/ng.2202.html#supplementary-information (2012).
Mangiarini, L. et al. Exon 1 of the HD Gene with an Expanded CAG Repeat Is Sufficient to Cause a Progressive Neurological Phenotype in Transgenic Mice. Cell 87, 493–506, doi: http://dx.doi.org/10.1016/S0092-8674(00)81369-0 (1996).
Cowin, R.-M. et al. Onset and Progression of Behavioral and Molecular Phenotypes in a Novel Congenic R6/2 Line Exhibiting Intergenerational CAG Repeat Stability. PLoS ONE 6, e28409, doi: 10.1371/journal.pone.0028409 (2011).
Morton, A. J. et al. Paradoxical delay in the onset of disease caused by super-long CAG repeat expansions in R6/2 mice. Neurobiol. Dis. 33, 331–341, doi: http://dx.doi.org/10.1016/j.nbd.2008.11.015 (2009).
Duzdevich, D. et al. Unusual Structures Are Present in DNA Fragments Containing Super-Long Huntingtin CAG Repeats. PLoS ONE 6, e17119, doi: 10.1371/journal.pone.0017119 (2011).
Dragatsis, I. et al. CAG repeat lengths ≥ 335 attenuate the phenotype in the R6/2 Huntington’s disease transgenic mouse. Neurobiol. Dis. 33, 315–330, doi: http://dx.doi.org/10.1016/j.nbd.2008.10.009 (2009).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, e45 (2001).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotech. 29, 644–652, doi: http://www.nature.com/nbt/journal/v29/n7/abs/nbt.1883.html#supplementary-information (2011).
Langfelder, P. et al. Integrated genomics and proteomics define huntingtin CAG length-dependent networks in mice. Nat. Neurosci. 19, 623–633, doi: 10.1038/nn.4256 (2016).
Wheeler, V. C. et al. Length-dependent gametic CAG repeat instability in the Huntington’s disease knock-in mouse. Hum. Mol. Genet. 8, 115–122 (1999).
Menalled, L. B. et al. Comprehensive Behavioral and Molecular Characterization of a New Knock-In Mouse Model of Huntington’s Disease: zQ175. PLoS ONE 7, e49838, doi: 10.1371/journal.pone.0049838 (2012).
Menalled, L. B., Sison, J. D., Dragatsis, I., Zeitlin, S. & Chesselet, M. F. Time course of early motor and neuropathological anomalies in a knock-in mouse model of Huntington’s disease with 140 CAG repeats. J. Comp. Neurol. 465, 11–26, doi: 10.1002/cne.10776 (2003).
Brooks, S., Higgs, G., Jones, L. & Dunnett, S. B. Longitudinal analysis of the behavioural phenotype in Hdh(CAG)150 Huntington’s disease knock-in mice. Brain Res. Bull. 88, 182–188, doi: 10.1016/j.brainresbull.2010.05.004 (2012).
Brooks, S., Higgs, G., Jones, L. & Dunnett, S. B. Longitudinal analysis of the behavioural phenotype in HdhQ92 Huntington’s disease knock-in mice. Brain Res. Bull. 88, 148–155, doi: 10.1016/j.brainresbull.2010.05.003 (2012).
Menalled, L. et al. Systematic behavioral evaluation of Huntington’s disease transgenic and knock-in mouse models. Neurobiol. Dis. 35, 319–336, doi: 10.1016/j.nbd.2009.05.007 (2009).
Wheeler, V. C. et al. Early phenotypes that presage late-onset neurodegenerative disease allow testing of modifiers in Hdh CAG knock-in mice. Hum. Mol. Genet. 11, 633–640 (2002).
Labadorf, A. et al. RNA Sequence Analysis of Human Huntington Disease Brain Reveals an Extensive Increase in Inflammatory and Developmental Gene Expression. PLoS ONE 10, e0143563, doi: 10.1371/journal.pone.0143563 (2015).
Jeong, Y. J. et al. 1950 MHz Electromagnetic Fields Ameliorate Abeta Pathology in Alzheimer’s Disease Mice. Curr. Alzheimer Res. 12, 481–492 (2015).
Olah, J. et al. Interactions of pathological hallmark proteins: tubulin polymerization promoting protein/p25, beta-amyloid, and alpha-synuclein. J. Biol. Chem. 286, 34088–34100, doi: 10.1074/jbc.M111.243907 (2011).
Benn, C. L., Fox, H. & Bates, G. P. Optimisation of region-specific reference gene selection and relative gene expression analysis methods for pre-clinical trials of Huntington’s disease. Mol. Neurodegener. 3, 17, doi: 10.1186/1750-1326-3-17 (2008).
Levin, J. Z. et al. Comprehensive comparative analysis of strand-specific RNA sequencing methods. Nat. Methods 7, 709–715, doi: 10.1038/nmeth.1491 (2010).
Parkhomchuk, D. et al. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 37, e123, doi: 10.1093/nar/gkp596 (2009).
Zhong, S. et al. High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harbor Protoc. 2011, 940–949, doi: 10.1101/pdb.prot5652 (2011).
Sugathan, A. et al. CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors. Proc. Natl. Acad. Sci. USA 111, E4468–E4477, doi: 10.1073/pnas.1405266111 (2014).
Andrews, S. FastQC A Quality Control tool for High Throughput Sequence Data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/, doi: citeulike-article-id:11583827.
Wu, T. D. & Nacu, S. Fast and SNP-tolerant detection of complex variants and splicing in short reads. Bioinformatics (Oxford, England) 26, 873–881, doi: 10.1093/bioinformatics/btq057 (2010).
DeLuca, D. S. et al. RNA-SeQC: RNA-seq metrics for quality control and process optimization. Bioinformatics (Oxford, England) 28, 1530–1532, doi: 10.1093/bioinformatics/bts196 (2012).
Wang, L., Wang, S. & Li, W. RSeQC: quality control of RNA-seq experiments. Bioinformatics (Oxford, England) 28, 2184–2185, doi: 10.1093/bioinformatics/bts356 (2012).
Li, H. & Handsaker, B. et al. The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics (Oxford, England), 25, 2078–2079, doi: 10.1093/bioinformatics/btp352 (2009).
Grabherr, M. G. et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652, doi: 10.1038/nbt.1883 (2011).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics (Oxford, England) 26, 841–842, doi: 10.1093/bioinformatics/btq033 (2010).
Blumenthal, I. et al. Transcriptional consequences of 16p11.2 deletion and duplication in mouse cortex and multiplex autism families. Am. J. Hum. Genet. 94, 870–883, doi: 10.1016/j.ajhg.2014.05.004 (2014).
Huang da, W., Sherman, B. T. & Lempicki, R. A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 4, 44–57, doi: 10.1038/nprot.2008.211 (2009).
Robinson, J. T. et al. Integrative genomics viewer. Nat. Biotechnol. 29, 24–26, doi: 10.1038/nbt.1754 (2011).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 29, 15–21, doi: 10.1093/bioinformatics/bts635 (2013).
Anders, S., Pyl, P. T. & Huber, W. HTSeq – A Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England), doi: 10.1093/bioinformatics/btu638 (2014).
Pollard, K. S., Hubisz, M. J., Rosenbloom, K. R. & Siepel, A. Detection of nonneutral substitution rates on mammalian phylogenies. Genome Res. 20, 110–121, doi: 10.1101/gr.097857.109 (2010).
Acknowledgements
The authors wish to thank Kristine Boxen of the Genomics Centre, Auckland Science Analytical Services, The University of Auckland, Auckland, New Zealand for assistance with Sanger sequencing services. This work was supported by the National Institutes of Health HD065286 (JFG), MH095867 (MET), GM061354 (MET, JFG), CHDI Inc. (JFG, AJM), NARSAD (MET), the Canada Research Chairs program and a grant from the Natural Science and Engineering Research Council of Canada (CE), and the Neurological Foundation of New Zealand (JJ).
Author information
Authors and Affiliations
Contributions
J.C.J., S.E., M.E.T. designed and conducted experiments and wrote the manuscript, C.C., C.H., R.R.H., D.D.B., A.S., and I.B. conducted experiments, S.J.R., R.G.S., M.E.M., C.E. and J.F.G. helped design experiments and A.J.M. provided mice. All authors reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Jacobsen, J., Erdin, S., Chiang, C. et al. Potential molecular consequences of transgene integration: The R6/2 mouse example. Sci Rep 7, 41120 (2017). https://doi.org/10.1038/srep41120
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep41120
This article is cited by
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.