Abstract
Adenylyl cyclase (AC), which produces the signalling molecule cAMP, has numerous important cellular functions in diverse organisms from prokaryotes to eukaryotes. Here we report the identification and characterization of an AC gene from the liverwort Marchantia polymorpha. The encoded protein has both a C-terminal AC catalytic domain similar to those of class III ACs and an N-terminal cyclic nucleotide phosphodiesterase (PDE) domain that degrades cyclic nucleotides, thus we designated the gene MpCAPE (COMBINED AC with PDE). Biochemical analyses of recombinant proteins showed that MpCAPE has both AC and PDE activities. In MpCAPE-promoter-GUS lines, GUS activity was specifically detected in the male sexual organ, the antheridium, suggesting MpCAPE and thus cAMP signalling may be involved in the male reproductive process. CAPE orthologues are distributed only in basal land plants and charophytes that use motile sperm as the male gamete. CAPE is a subclass of class III AC and may be important in male organ and cell development in basal plants.
Similar content being viewed by others
Introduction
Cyclic AMP (cAMP) is a second messenger controlling many cellular functions and is synthesized by adenylyl cyclases (ACs). The intracellular concentration of cAMP is tightly regulated by the activities of AC and its degradation enzyme, cAMP phosphodiesterase (PDE)1,2.
Genes encoding adenylyl cyclases have been isolated from most of species of organisms and the physiological functions of cAMP have been well characterized3,4,5,6. For example, in Escherichia coli, cAMP binds to a receptor protein called the cAMP receptor protein (CRP) and the cAMP-CRP complex regulates the transcriptional activation of catabolite-sensitive operons7. In mammals, an increased cAMP level in response to the actions of hormones such as glucagon and adrenaline promotes phosphorylation of several intracellular enzymes via activation of protein kinase A, resulting in the enhancement of enzyme activities in glycogen and lipid metabolisms8,9. cAMP also plays roles in learning, memory and olfactory sensation by regulating gene expression and channel activity10,11. In mammals, ACs are classified into nine transmembrane enzymes (tmACs) and one soluble enzyme (sAC)12. The sAC appears evolutionarily distinct from tmACs and more closely related to cyanobacterial ACs, and plays central roles in sperm capacitation13,14,15.
Much effort has been put into isolating an AC from plants because of its crucial functions in other organisms16,17,18. There are three reports of the identification of AC genes in plants: PSiP in Zea mays19, HpAC1 in Hippeastrum x hybridum20 and AtKUP7 in Arabidopsis thaliana21. However, the amino acid sequence of PSiP did not show any homology to known ACs. Furthermore, AtTTM3, the product of the HpAC1 orthologue in Arabidopsis thaliana, is not an AC and has tripolyphosphatase activity22. Because the four classes of ACs identified show no sequence homology among them, indicating that they have emerged independently by convergent evolution1,23, it is possible that a new class of AC might exist in land plants. This may be why it has been difficult to identify an AC in plants, especially in angiosperms, despite the availability of huge amounts of genomic information. The genomic information on basal land plants is rapidly increasing through the progress of their genome projects24,25,26. Thus, now is a good time to search for AC genes in the genomes of basal land plants.
In this report, we searched for AC genes in the transcriptome data of the liverwort Marchantia polymorpha and found one sequence that had significant similarity to class III ACs (the universal class comprising ACs from bacteria to mammals). In addition to the AC domain in the C-terminal part, the N-terminal part showed similarity to cyclic nucleotide phosphodiesterases, so the gene was designated COMBINED AC with PDE; CAPE. MpCAPE has Mn2+-enhanced adenylyl cyclase activity and thus is the first functional class III AC from land plants. Moreover, promoter-GUS analysis showed MpCAPE is specifically expressed in the antheridium. The distribution of CAPE orthologues corresponds to the streptophyte lineage that uses motile sperm as the male gamete for sexual reproduction. We discuss the involvement of CAPE in male reproductive organ development in streptophytes that have CAPE.
Results
Identification of an AC gene in M. polymorpha
A BLAST search was carried out against the transcriptome database of M. polymorpha to find genes encoding AC using the amino acid sequence of a cyanobacterial AC, CyaC27. We found one sequence showing significant similarity to the catalytic domain of CyaC. In addition to the AC domain, its deduced amino acid sequence contained a consensus sequence of the catalytic domain of PDEs, which are the degradation enzymes of the cyclic nucleotides, cAMP and cyclic GMP (cGMP). Thus, we designated the gene MpCAPE (COMBINED AC with PDE) according to the gene nomenclature of Marchantia28. MpCAPE is registered as Mapoly0068s0004 in the Phytozome web site (https://phytozome.jgi.doe.gov).
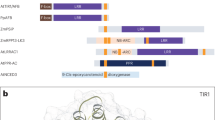
Reverse transcription (RT)-PCR was performed using total RNA from M. polymorpha and the amino acid sequence of MpCAPE was determined (Supplementary Fig. S1). MpCAPE consisted of 1610 amino acids with a calculated molecular mass of 179 kDa. The C- and N-terminal parts showed similarity to the catalytic domains of class III ACs and PDEs, respectively (Fig. 1). Although the sequence identity was low, the consensus amino acids required for catalysis and substrate binding29,30 were well conserved in the AC domain of MpCAPE (Fig. 1B, asterisks). In PDEs, two metal ions, Zn2+ and Mg2+, are coordinated by highly conserved amino acids in the catalytic domain and indispensable for catalysis31,32. In the PDE domain of MpCAPE, all amino acids required for metal binding were conserved (Fig. 1C, red asterisks). In the middle part of MpCAPE, there were two hydrophobic segments, which were predicted to act as transmembrane helices (Fig. 1A and Supplementary Fig. S1).
Domain organization and amino acid alignment of the AC and PDE domains of MpCAPE.
(A) Schematic representation of the full-length MpCAPE. The catalytic domains of AC and PDE are shown as red and green boxes, respectively. Membrane-spanning regions are shown as blue boxes (TM1 and TM2). (B) An alignment of the AC domain of MpCAPE with Arthrospira platensis CyaC (ApCyaC), Sinorhizobium meliloti CyaA (SmCyaA), Euglena gracilis PACa (EgPACaC1 and EgPACaC2), Bos taurus type 1 AC (BtACt1C1 and BtACt2C2) and Homo sapiens type 10 AC (HsACt10C1 and HsACt10C2). Amino acids involved in binding the substrate ATP are indicated by red asterisks. (C) An alignment of the PDE domain of MpCAPE with Homo sapiens PDE4B, 8 A and 9 A. Amino acids involved in metal ion binding are indicated by red asterisks. Amino acid residues identical in majority of the sequences are shown in black.
Complementation of the AC-deficient (∆cya) E. coli strain MK1010
Wild-type E. coli cells utilize maltose in the presence of intracellular cAMP and form red colonies on MacConkey-maltose agar plates because of acid production during their growth, whereas ∆cya mutants, including the E. coli MK1010 strain, form white colonies33. To examine whether MpCAPE has cAMP production activity, the expression vector pGEX-MpCAPE-AC, which contained the sequence of the putative AC domain of MpCAPE (1251–1610), was constructed and introduced into E. coli MK1010. The transformant pGEX-MpCAPE-AC/MK1010 formed red colonies (Fig. 2), similar to the transformant pGEX-CyaG-CD/MK1010, in which the catalytic domain of the cyanobacterial adenylyl cyclase CyaG was expressed34, whereas the transformant pGEX-6P-1/MK1010 (vector control) formed white colonies (Fig. 2). Thus, pGEX-MpCAPE-AC complemented the AC-deficient phenotype of E. coli MK1010. Additionally, pGEX-MpCAPE-AC(D1340A) was constructed because Asp-1340 corresponds to the metal binding site essential for the catalytic activity of mammalian AC35. The point mutation (D1340A) prevented pGEX-MpCAPE-AC from complementing the AC deficiency of E. coli MK1010 (Fig. 2). These results suggested that the AC domain of MpCAPE had an AC activity.
Detection of cAMP in E. coli MK1010 harboring a gene for the AC of MpCAPE
The cellular cAMP levels in E. coli MK1010-based transformants were measured (Table 1). cAMP was not detected in E. coli MK1010, even when pGEX-6P-1 was introduced. In contrast, cAMP was detected in E. coli MK1010 transformed with pGEX-MpCAPE-AC, as was the case with the wild-type (cya+) strain E. coli DH5α. The D1340A mutation in MpCAPE-AC caused the loss of the ability to produce cAMP.
In vitro AC activity of GST-MpCAPE-AC
To analyze its AC activity in vitro, the catalytic domain of MpCAPE-AC was produced as a GST fusion protein (GST-MpCAPE-AC) in E. coli and purified (Supplementary Fig. S2). The specific activity of GST-MpCAPE-AC with Mn2+ was 35-fold higher than that with Mg2+ (Table 2). The enhancement of the AC activity by Mn2+ is similar to other class III ACs14,27,36,37. The results of mutation analysis using GST-MpCAPE-AC(D1340A) showed that Asp-1340 was essential for AC activity (Table 2). Bicarbonate stimulates the activities of mammalian sAC and a cyanobacterial AC15. We examined the effect of bicarbonate on the AC activity of GST-MpCAPE-AC under the basal condition of its activity in the presence of Mg2+. The AC activity of GST-MpCAPE-AC was not affected by bicarbonate (Supplementary Table S1). The guanylyl cyclase (GC) activity of GST-MpCAPE-AC was tested by adding GTP instead of ATP to the enzyme assay mixtures but no GC activity was detected.
In vitro PDE activity of His-MpCAPE-PDE
To analyze its PDE activity in vitro, the catalytic domain of MpCAPE-PDE was produced as a 6 × His fusion protein (His-MpCAPE-PDE) in E. coli and purified using Ni2+-Sepharose column. The His-MpCAPE-PDE was eluted from the column with several other proteins (Supplementary Fig. S3A, lane 1). Using the partially purified protein sample, the PDE activity of His-MpCAPE-PDE was assayed in the presence or absence of divalent cations (Fig. 3). The result showed that His-MpCAPE-PDE hydrolyzed both cAMP and cGMP but cAMP was much more favorable substrate than cGMP. Divalent cations (Mg2+, Mn2+ and Fe2+) stimulated both cAMP- and cGMP-dependent activities. PDE activities in the presence of Mg2+ were examined using mutant proteins, His-MpCAPE-PDE-H199Q and His-MpCAPE-PDE-H203Q (Supplementary Fig. S3), in which the highly conserved histidines (Fig. 1C) were replaced by glutamine. PDE activities were completely disappeared by the mutations (Table S2). The result suggested that His-199 and His-203 were essential for the catalytic activity of MpCAPE-PDE and the activity detected in the partially purified sample was not due to the contamination proteins from E. coli.
Effect of divalent cations on phosphodiesterase activity of His-MpCAPE-PDE.
Phosphodiesterase activity of partially purified His-MpCAPE-PDE protein sample (13 μg) was measured in the presence or absence of various cations. cAMP or cGMP was used as substrate. The inset shows the magnification of the data with cGMP. Values indicate means ± SD (n = 3). n.d.: not detected.
Tissue-specific expression pattern of MpCAPE
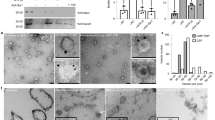
To examine the developmental and tissue-specific MpCAPE expression, mRNA accumulation was examined by RT-PCR. The result showed that MpCAPE specifically expressed in the antheridiophore, which is the male gametophore bearing the sexual organ, the antheridium (Fig. 4A). Next, we generated transgenic M. polymorpha lines expressing the GUS gene under the control of the MpCAPE promoter. No GUS expression was detected in the vegetative growth phase (Fig. 4B–E). GUS expression was observed as dots in a sample of the antheridiophore (Fig. 4F). The GUS-stained dots looked like antheridia, so a GUS-stained antheridiophore was dissected and antheridia were prepared. GUS expression was observed in the antheridium (Fig. 4H). GUS expression was not detected in the female gametophore, the archegoniophore (Fig. 4G).
MpCAPE mRNA accumulation in wild type and MpCAPE promoter-GUS expression pattern in transgenic M. polymorpha plants.
(A) RT-PCR analysis of CAPE mRNA accumulation in antheridiophores (lane 1), archegoniophores (lane 2), thalli from male accession Tak-1 (lane 3) and thalli from female accession Tak-2 (lane 4). EF1α encoding elongation factor 1α was used as a control. GUS expression in gemmalings (B and C), thalli (D and E), antheridiophore (F) and archegoniophore (G) of MpCAPE promoter-GUS transgenic plants. Backgrounds of the transgenic plants were Tak-1 (B, D and F) and Tak-2 (C, E and G). GUS activity was detected only in the antheridiophore. Bars = 1 mm. (H) Antheridium dissected from a GUS-stained antheridiophore. Bar = 0.1 mm.
CAPE orthologues in Streptophyta
We detected orthologous sequences in the moss Physcomitrella patens and the lycophyte Selaginella moellendorffii from their complete genome databases and in the charophyte Coleochaete orbicularis from its transcriptome data38 (Supplementary Fig. S4). Moreover, cDNA fragments of CAPE were amplified from the charophyte Chara braunii and the pteridophyte Adiantum capillus-veneris by RT-PCR, and their amino acid sequences were deduced (Supplementary Fig. S4). However, we could not find CAPE orthologues in gymnosperms (Picea abies and Pinus taeda) and angiosperms. Several homologous sequences that contained an AC domain but not a PDE domain were also found in green algae including the charophytes Mesostigma viride and Klebsormidium flaccidum from their transcriptome and complete genome databases38,39, respectively, and chlorophytes such as Chlamydomonas reinhardtii, Coccomyxa subellipsoidea, Ostreococcus tauri and Micromonas pusilla. Phylogenetic analysis showed that, regarding their AC domains, CAPEs and algal AC-like sequences were separated into two clades, with the branch supported by a high bootstrap value (72, Fig. 5).
Phylogenetic tree of ACs.
The phylogenetic tree was inferred using the maximum-likelihood method with the LG+Gamma model. Numbers represent support values (>50%) obtained with 100 bootstrap replicates using the MEGA7 software (LG+Gamma mode). The evolutionary distances were computed in units of the number of amino acid substitutions per site, as shown by the scale bar below the tree. The accession numbers of AC sequences used for phylogenetic analysis are shown in Supplementary Table S3.
Discussion
We found a unique AC with a PDE domain, CAPE, in the genome of M. polymorpha and characterized the AC activity of MpCAPE using an E. coli ∆cya mutant and recombinant proteins. From the following results, we conclude that the protein encoded by MpCAPE of M. polymorpha can produce cAMP and is the first functional class III AC identified from land plants. (i) The amino acid sequence deduced from the isolated cDNA exhibited similarity to the catalytic domain of class III ACs. Importantly, the amino acid residues required for the catalytic activity were conserved (Fig. 1). (ii) The cDNA fragment encoding the AC domain of MpCAPE complemented ∆cya of the E. coli MK1010 mutant strain (Fig. 2). (iii) cAMP was detected in the E. coli MK1010 strain transformed with the AC fragment of MpCAPE (Table 1). (iv) A recombinant protein consisting of the AC domain of MpCAPE produced cAMP in vitro (Table 2).
The AC activity of MpCAPE was enhanced by Mn2+. Mn2+-enhancement has been observed in other ACs14,27,36,37 and is a common property in class III ACs. Thus, in the conservation of its amino acid sequence and its enzymatic properties, MpCAPE retains the characteristics of class III ACs. Furthermore, an aspartate residue, corresponding to Asp-1340 in MpCAPE, is essential for ATP binding by associating with Mg2+ in class III ACs29,40. To confirm that MpCAPE produces cAMP, an AC mutant in which Asp-1340 was replaced with Ala was tested for AC activities, i.e., complementation of the E. coli ∆cya mutant, cAMP production in E. coli cells and in vitro AC activity. In all experiments, the mutation (D1340A) resulted in the disappearance of AC activity, suggesting that MpCAPE certainly encodes an AC.
The AC activities of MpCAPE with Mn2+ were 7 times higher than that of AtKUP7 of Arabidopsis thaliana (2.2 pmol min−1 mg−1 with Mn2+)21. Those proteins derived from land plants seem to have an equivalent AC activity. On the other hand, mammalian ACs show much higher activities, for example, 75 pmol min−1 mg−1 for a transmembrane AC (AC5) and 10 nmol min−1 mg−1 for soluble AC (sAC)15. It is possible that cAMP effectors might be localized in close proximity to ACs and relatively low AC activity might be enough for their activation. Also, there might be a mechanism to stimulate AC activity in plant cells.
MpCAPE is encoded by a single-copy gene and differs from the protein encoded by CUFF.20439 (Mapoly0178s0022) that was annotated as an AC gene in Higo et al.41. The Mapoly0178s0022 protein does not contain a PDE domain but does have a putative AC catalytic domain on its N-terminal side in which several conserved amino acids for catalysis are missing. Nevertheless, the enzymatic activity of the Mapoly0178s0022 protein needs to be characterized because it might still have the activity of an AC or GC, whose catalytic core sequence is homologous to that of AC35.
MpCAPE may be a membrane protein and has two-membrane-spanning helices between the AC and PDE domains, so both domains may face on the same side of a membrane, likely the cytosolic space because of the presence of ATP pool as substrate for the AC activity. The cellular level of cAMP could be tightly controlled by MpCAPE through synthesis and hydrolysis. It is thought that, because there are different cAMP effectors that regulate each specific signalling process in a cell, cAMP must be prevented from free diffusion and localized in restricted areas to activate specific signalling pathways42. PDEs have a critical role in the compartmentalization of cAMP signalling43,44. MpCAPE allows for such spatial regulation of cAMP by itself.
The gametophyte of M. polymorpha is dioecious. When entering the sexual reproductive phase, the male and female gametophytes develop individual sexual organs, the antheridium and archegonium, on special gametophores called the antheridiophore and archegoniophore, respectively45. It has been shown that a number of specific genes are expressed in the antheridium of M. polymorpha41. In our MpCAPE promoter-GUS experiment, the promoter activity was specifically detected in the antheridium (Fig. 4). Motile sperm cells with two flagella are developed in the antheridium of M. polymorpha 45. As far as we know, there have been no reports showing a role for cAMP in antheridium formation or spermatogenesis in M. polymorpha. However, in animal cells including mammalian cells, the function of cAMP in spermatogenesis has been characterized and it has been shown to be an indispensable factor in sperm physiology, such as the regulation of sperm capacitation, the acrosome reaction and the activation of sperm motility13,46,47,48,49. In addition to the physiological roles of cAMP, the ACs that play roles in spermatogenesis have been analyzed13,50. Mammalian ACs include nine transmembrane AC isoforms (tmAC1–9) and one soluble AC (sAC)51. The sAC not only has a different topology to tmACs, but also a distinct catalytic domain, which is more closely related to cyanobacterial adenylyl cyclases14,15. In mammalian sperms, the sAC is the main source of cAMP and has a dominant role in the acquisition of fertilizing capacity13. The fact that AC has a dominant function in sperm physiology and has been genetically inherited during the evolution of mammals may suggest a possible role for cAMP in the reproductive organ development of M. polymorpha. A recent paper has shown the expression of a cAMP-dependent protein kinase and cyclic nucleotide-gated ion channel in the antheridium of M. polymorpha41. It is likely that these proteins function as signalling factors downstream of MpCAPE.
The functional importance of the MpCAPE in the male organ also seems to be supported by the distribution of CAPEs in Streptophyta; charophytes plus land plants. In charophytes, CAPE genes were identified in Chara and Coleochaete, but not in the genome of Klebsormidium, although an algal AC-like sequence is present in its genome. According to the phylogeny of the charophyte lineage52,53, since Charales appeared and diverged after the establishment of Mesostigmatales and Klebsormidiales, CAPE must have appeared during the evolutionary process between Klebsormidiales and Charales. Charales first developed a motile sperm with flagella in Streptophyta and the architecture of the mature sperm is remarkably similar to that in basal land plants54. The occurrence of CAPE is in consistent with the emergence of motile sperm as the male gamete in Charales. Furthermore, because the AC domains of CAPEs were sister to the algal AC-like sequences of Mesostigma, Klebsormidium and chlorophytes in the phylogenetic analysis of class III ACs (Fig. 5), we can infer that CAPEs arose from the fusion of a PDE domain with an algal AC-like sequence.
Zygnematophyceae, a class of charophytes, have been proposed as the sister group of land plants, but do not produce motile sperm cells and alternatively use conjugation system for sexual reproduction52,53. We could not find homologous sequences of CAPE in the transcriptome data of Spirogyra pratensis38 and in the GenBank sequence data derived from the members of Zygnematophyceae. Complete genome sequence data from Zygnematophyceae will reveal the presence or absence of CAPE in this lineage.
In land plants, CAPEs are present in Marchantia, Physcomitrella, Selaginella and Adiantum, which all use motile sperm as male gametes54. However, neither CAPEs nor class III ACs including algal AC-like sequences are present in gymnosperms (Picea abies and Pinus taeda) and angiosperms that use a non-motile sperm cell delivered to the egg cell. CAPE seems to have been lost during the evolutionary process from ferns to seed plants. The disappearance of CAPEs in land plant lineages also corresponds with the loss of motile sperm. In summary, the distribution of CAPEs coincides well with the use of motile sperm in Streptophyta. It will be interesting to investigate whether CAPEs exist in the gymnosperms Ginkgo and Cycas, which are unique in having motile sperms as male gametes.
Using recently developed molecular techniques for M. polymorpha55, analysis of MpCAPE mutants constructed by gene targeting should clarify the physiological roles of cAMP in the male organ of M. polymorpha. Information on the physiological and molecular functions of MpCAPE will contribute to our understanding of the role of cAMP in other plants.
Methods
Culture and growth conditions of Marchantia polymorpha
Male and female accessions of M. polymorpha, Takaragaike (Tak)-1 and Tak-2, respectively, were cultured aseptically at 22 °C on 1/2-strength Gamborg’s B5 agar medium under continuous white light conditions, or grown on vermiculite soaked with 1/1000-diluted Hyponex solution (HYPONex JAPAN Co. Ltd, Osaka, Japan) at 22 °C under 16 h white fluorescent light supplemented with far-red LED light and 8 h dark conditions.
Cloning of genes encoding an AC from M. polymorpha
A MpCAPE cDNA was obtained by RT-PCR using total RNA prepared from antheridiophores of M. polymorpha Tak-1 and the primers MpCAPE-f (5′-CACCATGCATGCTTGCTTTGAGGG-3′) and MpCAPE-r (5′-CTACTTCTCGGTGAGTTCTC-3′). The amplified DNA fragment (approximately 5 kb) was excised from an agarose gel and purified using a NucleoSpin Extract kit (Macherey-Nagel, Germany). The purified DNA fragment was cloned into the cloning vector pENTR/D-TOPO (Thermo Fisher Scientific, USA). The nucleotide sequence was determined using a DNA sequencer (Genetic Analyzer 3130, Thermo Fisher Scientific, USA).
Construction of the expression plasmid for the GST-MpCAPE-AC protein
A DNA fragment encoding the AC domain of MpCAPE (1251–1610) was amplified by PCR using the primers MpCAPE-EX-f (5′-GGGATCCCCGGAATTCCAGCCTATTGAGCGCATGGT-3′) and MpCAPE-EX-r (5′-AGTCACGATGCGGCCGCCTACTTCTCGGTGAGTTCTC-3′), and cDNA as a template. The amplified DNA was cloned into the EcoRI-NotI site of the pGEX-6P-1 vector with an In-fusion Cloning kit (Takara, Japan). The resulting plasmid was named pGEX-MpCAPE-AC.
The GST-MpCAPE-AC(D1340A) mutant was constructed with mismatched oligonucleotides and PCR. PCRs were performed with the primers MpCAPE-EX-f and MpAC-D2A-r (5′-AGTTTCGTATGgCACAGAATCCA-3′) or MpAC-D2A-f (5′-TGGATTCTGTGcCATACGAAACT-3′) and MpCAPE-EX-r. The resulting amplified DNA fragments were mixed and PCR was repeated with the primers MpCAPE-EX-f and MpCAPE-EX-r using the DNA mixture as a template. The amplified DNA fragment was cloned into the EcoRI-NotI site of the pGEX-6P-1 vector as described above. The resulting plasmid was named pGEX-MpCAPE-AC(D1340A).
Expression and purification of GST-MpCAPE-AC proteins
The constructed expression plasmids were introduced into an E. coli AC mutant, MK101033, to express the AC domain of MpCAPE as a fusion protein with an affinity tag, glutathione-S-transferase (GST). The transformants were grown at 25 °C in LB medium (1.5 L) containing ampicillin (100 μg ml−1) and kanamycin (50 μg ml−1). Protein expression was induced by adding 0.1 mM isopropyl-ß-D-thiogalactopyranoside (IPTG) at OD600 = 0.5. The cells were grown at 25 °C for 24 h, harvested by centrifugation, resuspended in 20 mL of TEG buffer (50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 10% (w/v) glycerol, 0.5 M NaCl) and disrupted by sonication. The cell extracts were centrifuged at 15,000 × g for 30 min and the supernatants were loaded onto a 1 mL glutathione column (GSTrap HP, GE Healthcare, USA). The columns were washed with TEG buffer and the proteins were eluted with 5 mM glutathione in TEG buffer.
Adenylyl cyclase activity assay
In vitro adenylyl cyclase reactions (9 μg of protein) were performed in 0.1 mL assay buffer containing 50 mM Tris-HCl (pH 7.5), 1 mM ATP, 1 mM DTT, and 1 mM MgCl2 or MnCl2 at 37 °C for 30 min. The enzyme reaction was terminated by adding 1 mL of 5% (v/v) trichloroacetic acid (TCA). After removing the TCA from each reaction mixture by extracting with ethyl ether, the samples were lyophilized. cAMP contents were measured with an enzyme immuno assay system (cAMP EIA system, GE Healthcare, USA) according to the manufacturer’s instructions.
Complementation test of the adenylyl cyclase deficiency of E. coli MK1010
The transformants (MK1010 cells harboring the constructed vectors described above) were streaked onto MacConkey agar plates (Difco, Germany) containing 1% (w/v) maltose, 100 μg ml−1 ampicillin and 50 μg ml−1 kanamycin and grown at 25 °C.
Construction of the expression plasmids for His-MpCAPE-PDE proteins
A DNA fragment encoding the PDE domain of MpCAPE (101–479) was amplified by PCR using the primers MpCAPE-PDE-EX-f (5′-TCGCGGATCCGAATTCCAGGGAATTAACTCGTGGAC-3′) and MpCAPE-PDE-EX-r (5′-GGTGGTGGTGCTCGAGTTAAAAGGGACCAAGAATCTGCT-3′), and cDNA as a template. The amplified DNA was cloned into the EcoRI-XhoI site of the pET28a vector with an In-fusion Cloning kit (Takara, Japan). The resulting plasmid was named pET-MpCAPE-PDE.
The His-MpCAPE-PDE(H199Q) and –PDE(H203Q) mutants were constructed with mismatched oligonucleotides and PCR in the same manner as described above for the construction of GST-MpCAPE-AC(D1340A) mutant. The amplified DNA fragments were cloned into the EcoRI-XhoI site of the pET28a vector. The resulting plasmids were named pET-MpCAPE-PDE(H199Q) and pET-MpCAPE-PDE(H203Q).
Expression and purification of His-MpCAPE-PDE proteins
The constructed expression plasmids were introduced into an E. coli Rosetta2(DE3)pLysS strain, to express the PDE domain of MpCAPE as a fusion protein with an affinity tag (6 ×His) and an epitope tag (T7-tag). The transformants were grown at 25 °C in LB medium (4 L) containing kanamycin (50 μg ml−1) and chloramphenicol (30 μg ml−1). Protein expression was induced by adding 0.1 mM IPTG at OD600 = 0.35. The cells were grown at 25 °C for 7 h, harvested by centrifugation, resuspended in 70 mL of TNG buffer (50 mM Tris-HCl (pH 8.0), 0.2 M NaCl, 10% (w/v) glycerol) and disrupted by sonication. The cell extracts were centrifuged at 15,000 × g for 30 min and the supernatants were loaded onto a 1 mL Ni2+ column (HisTrap HP, GE Healthcare, USA). The columns were washed with TNG buffer containing 100 mM imidazole and the proteins were eluted with 200 mM imidazole in TNG buffer.
Phosphodiesterase activity assay
In vitro phosphodiesterase reactions (13 μg of partially purified protein) were performed in 0.1 mL assay buffer containing 30 mM Tris-HCl (pH 8.0), 0.5 mM cAMP or cGMP, 0.1% (v/v) 2-mercaptoethanol, and 0.5 mM MgCl2, MnCl2 or FeCl2 at 37 °C for 20 min. The enzyme reaction was terminated by incubation at 100 °C for 10 min. After centrifugation at 15,000× g for 10 min, the supernatants were analyzed by reverse-phase column chromatography (COSMOSIL 5C18-AR; 4.6 × 250 mm; Nacalai, Kyoto, Japan) using 30 mM sodium phosphate buffer (pH 5.0) containing 5%(v/v) acetonitrile as the mobile phase at a flow rate of 1.0 ml min−1. The effluent was monitored at 259 nm to detect AMP and GMP hydrolyzed from cAMP and cGMP by phosphodiesterase, respectively.
Promoter-GUS construct and GUS staining
An 11-kb genomic DNA fragment, upstream of the triplet encoding His-259 of MpCAPE, was amplified by PCR using the primers PMpCAPE-f (5′-CACCACCAGCTAGGGAAACAGGGT-3′) and PMpCAPE-r (5′-GTGTCTCTGCTCCTCTTCAC-3′) and genomic DNA of M. polymorpha Tak-1 as a template. The amplified DNA fragment was cloned into the cloning vector pENTR/D-TOPO (Thermo Fisher Scientific, USA). The resulting plasmid was used for LR recombination by the Gateway technique (Thermo Fisher Scientific, USA) with pMpGWB10456 containing a GUS gene to produce the binary vector pMpGWB-Pcape. The GUS protein should be expressed as a translational fusion with the N-terminal fragment of MpCAPE (Met-1 to His-259). Agrobacterium-mediated transformation was carried out using regenerating thalli of M. polymorpha Tak-1 and Tak-257. Hygromycin-resistant plantlets were selected to establish isogenic lines. Gemmae obtained from the isogenic lines were planted on vermiculite, grown for an appropriate length of time and histochemically stained to detect GUS activity.
Sequence determination of MpCAPE orthologues of C. braunii and A. capillus-veneris
Primer pairs, CbCAPE-F (5′-CACCACGGTCCTCGTCTCAGTGTT-3′) & CbCAPE-R (5′-CGTCGTGTGCAACCCTATTT-3′) and AcCAPE-F (5′-CACCCCAAAGATGGTGGAACGAAT-3′) & AcCAPE-R (5′-TGGCATACCAAAATTCCACA-3′) for amplification of C. braunii and A. capillus-veneris CAPE cDNAs, respectively, were designed from the RNA-seq data obtained with an Illumina Hiseq2000. RT-PCR was performed using total RNA prepared from thalli of C. braunii or prothallia of A. capillus-veneris and the primers noted above. The amplified DNA fragments were cloned into the cloning vector pENTR/D-TOPO (Thermo Fisher Scientific, USA). The nucleotide sequences were determined using a DNA sequencer (Genetic Analyzer 3130, Thermo Fisher Scientific, USA).
Phylogenetic analysis
The amino acid sequences of the catalytic domains of adenylyl cyclases (ACs) (Table S3) were aligned using the ClustalX 2.0 program58. After removing ambiguously aligned regions, phylogenetic analysis was performed with a data matrix consisting of 134 amino acids for the AC domains from 26 operational taxonomic units (OTUs). A maximum-likelihood (ML) tree for the AC domains was determined using the MEGA7 software59, based on the LG60 + Gamma model. Bootstrap analysis of the ML tree was performed with 100 replications.
Other analytical procedures
Protein content was measured with a Bio-Rad protein assay kit using gamma-globulin as the standard.
Additional Information
How to cite this article: Kasahara, M. et al. An adenylyl cyclase with a phosphodiesterase domain in basal plants with a motile sperm system. Sci. Rep. 6, 39232; doi: 10.1038/srep39232 (2016).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Danchin, A. Phylogeny of adenylyl cyclases. Adv. Second Messenger Phosphoprotein Res. 27, 109–162 (1993).
Conti, M. & Beavo, J. Biochemistry and physiology of cyclic nucleotide phosphodiesterases: essential components in cyclic nucleotide signaling. Annu. Rev. Biochem. 76, 481–511 (2007).
Botsford, J. L. & Harman, J. G. Cyclic AMP in prokaryotes. Microbiol. Rev. 56, 100–122 (1992).
Montminy, M. Transcriptional regulation by cyclic AMP. Annu. Rev. Biochem. 66, 807–822 (1997).
D’Souza, C. A. & Heitman, J. Conserved cAMP signaling cascades regulate fungal development and virulence. FEMS Microbiol. Rev. 25, 349–364 (2001).
Parent, C. A. & Devreotes, P. N. Molecular genetics of signal transduction in Dictyostelium. Annu. Rev. Biochem. 65, 411–440 (1996).
Ullmann, A. & Danchin, A. Role of Cyclic AMP in bacteria. Adv. Cyclic Nucleotide Res. 15, 1–53 (1983).
Carmen, G. Y. & Victor, S. M. Signalling mechanisms regulating lipolysis. Cell. Signal. 18, 401–408 (2006).
Cohen, P. Protein phosphorylation and the control of glycogen metabolism in skeletal muscle. Philos. Trans. R. Soc. Lond., B, Biol. Sci. 302, 13–25 (1983).
Hanoune, J. & Defer, N. Regulation and role of adenylyl cyclase isoforms. Annu. Rev. Pharmacol. Toxicol. 41, 145–174 (2001).
Zagotta, W. N. & Siegelbaum, S. A. Structure and function of cyclic nucleotide-gated channels. Annu. Rev. Neurosci. 19, 235–263 (1996).
Steegborn, C. Structure, mechanism, and regulation of soluble adenylyl cyclases - similarities and differences to transmembrane adenylyl cyclases. Biochim. Biophys. Acta 1842, 2535–2547 (2014).
Buffone, M. G., Wertheimer, E. V., Visconti, P. E. & Krapf, D. Central role of soluble adenylyl cyclase and cAMP in sperm physiology. Biochim. Biophys. Acta 1842, 2610–2620 (2014).
Buck, J., Sinclair, M. L., Schapal, L., Cann, M. J. & Levin, L. R. Cytosolic adenylyl cyclase defines a unique signaling molecule in mammals. Proc. Natl. Acad. Sci. USA 96, 79–84 (1999).
Chen, Y. et al. Soluble adenylyl cyclase as an evolutionarily conserved bicarbonate sensor. Science 289, 625–628 (2000).
Newton, R. P., Roef, L., Witters, E. & Van Onckelen, H. Cyclic nucleotides in higher plants: theenduring paradox. New Phytol. 143, 427–455 (1999).
Assmann, S. M. Cyclic AMP as a second messenger in higher plants (Status and Future Prospects). Plant Physiol. 108, 885–889 (1995).
Gehring, C. Adenyl cyclases and cAMP in plant signaling - past and present. Cell Commun. Signal 8, 15 (2010).
Moutinho, A., Hussey, P. J., Trewavas, A. J. & Malho, R. cAMP acts as a second messenger in pollen tube growth and reorientation. Proc. Natl. Acad. Sci. USA 98, 10481–10486 (2001).
Swiezawska, B. et al. Molecular cloning and characterization of a novel adenylyl cyclase gene, HpAC1, involved in stress signaling in Hippeastrum x hybridum. Plant Physiol. Biochem. 80, 41–52 (2014).
Al-Younis, I., Wong, A. & Gehring, C. The Arabidopsis thaliana K+-uptake permease 7 (AtKUP7) contains a functional cytosolic adenylate cyclase catalytic centre. FEBS Lett. 589, 3848–3852 (2015).
Moeder, W. et al. Crystal structure and biochemical analyses reveal that the Arabidopsis triphosphate tunnel metalloenzyme AtTTM3 is a tripolyphosphatase involved in root development. Plant J. 76, 615–626 (2013).
Sismeiro, O., Trotot, P., Biville, F., Vivares, C. & Danchin, A. Aeromonas hydrophila adenylyl cyclase 2: a new class of adenylyl cyclases with thermophilic properties and sequence similarities to proteins from hyperthermophilic archaebacteria. J. Bacteriol. 180, 3339–3344 (1998).
Rensing, S. A. et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319, 64–69 (2008).
Banks, J. A. et al. The Selaginella genome identifies genetic changes associated with the evolution of vascular plants. Science 332, 960–963 (2011).
Bowman, J. L., Araki, T. & Kohchi, T. Marchantia: past, present and future. Plant Cell Physiol. 57, 205–209 (2016).
Kasahara, M., Yashiro, K., Sakamoto, T. & Ohmori, M. The Spirulina platensis adenylate cyclase gene, cyaC, encodes a novel signal transduction protein. Plant Cell Physiol. 38, 828–836 (1997).
Bowman, J. L. et al. The Naming of Names: Guidelines for Gene Nomenclature in Marchantia. Plant Cell Physiol. 57, 257–261 (2016).
Hurley, J. H. Structure, mechanism, and regulation of mammalian adenylyl cyclase. J. Biol. Chem. 274, 7599–7602 (1999).
Linder, J. U. & Schultz, J. E. The class III adenylyl cyclases: multi-purpose signalling modules. Cell. Signal. 15, 1081–1089 (2003).
Francis, S. H., Colbran, J. L., McAllister-Lucas, L. M. & Corbin, J. D. Zinc interactions and conserved motifs of the cGMP-binding cGMP-specific phosphodiesterase suggest that it is a zinc hydrolase. J. Biol. Chem. 269, 22477–22480 (1994).
Xu, R. X. et al. Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity. Science 288, 1822–1825 (2000).
Kawamukai, M. et al. Nucleotide sequence and characterization of the sfs1 gene: sfs1 is involved in CRP*-dependent mal gene expression in Escherichia coli. J. Bacteriol. 173, 2644–2648 (1991).
Kasahara, M., Unno, T., Yashiro, K. & Ohmori, M. CyaG, a novel cyanobacterial adenylyl cyclase and a possible ancestor of mammalian guanylyl cyclases. J. Biol. Chem. 276, 10564–10569 (2001).
Liu, Y., Ruoho, A. E., Rao, V. D. & Hurley, J. H. Catalytic mechanism of the adenylyl and guanylyl cyclases: modeling and mutational analysis. Proc. Natl. Acad. Sci. USA 94, 13414–13419 (1997).
Londos, C. & Preston, M. S. Activation of the hepatic adenylate cyclase system by divalent cations. J. Biol. Chem. 252, 5957–5961 (1977).
Field, J. et al. Purification of a RAS-responsive adenylyl cyclase complex from Saccharomyces cerevisiae by use of an epitope addition method. Mol. Cell. Biol. 8, 2159–2165 (1988).
Ju, C. et al. Conservation of ethylene as a plant hormone over 450 million years of evolution. Nat. Plants 1, 14004 (2015).
Hori, K. et al. Klebsormidium flaccidum genome reveals primary factors for plant terrestrial adaptation. Nat. Commun 5, 3978 (2014).
Tesmer, J. J., Sunahara, R. K., Gilman, A. G. & Sprang, S. R. Crystal structure of the catalytic domains of adenylyl cyclase in a complex with Gsα-GTPγS. Science 278, 1907–1916 (1997).
Higo, A. et al. Transcriptional framework of male gametogenesis in the liverwort Marchantia polymorpha L. Plant Cell Physiol. 57, 325–338 (2016).
Houslay, M. D. & Adams, D. R. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 370, 1–18 (2003).
Baillie, G. S. Compartmentalized signalling: spatial regulation of cAMP by the action of compartmentalized phosphodiesterases. FEBS J. 276, 1790–1799 (2009).
Conti, M., Mika, D. & Richter, W. Cyclic AMP compartments and signaling specificity: role of cyclic nucleotide phosphodiesterases. J. Gen. Physiol. 143, 29–38 (2014).
Shimamura, M. Marchantia polymorpha: taxonomy, phylogeny and morphology of a model system. Plant Cell Physiol. 57, 230–256 (2016).
Inaba, K., Morisawa, S. & Morisawa, M. Proteasomes regulate the motility of salmonid fish sperm through modulation of cAMP-dependent phosphorylation of an outer arm dynein light chain. J. Cell Sci. 111, 1105–1115 (1998).
Izumi, H., Marian, T., Inaba, K., Oka, Y. & Morisawa, M. Membrane hyperpolarization by sperm-activating and -attracting factor increases cAMP level and activates sperm motility in the ascidian Ciona intestinalis. Dev. Biol. 213, 246–256 (1999).
Visconti, P. E. et al. Capacitation of mouse spermatozoa. II. Protein tyrosine phosphorylation and capacitation are regulated by a cAMP-dependent pathway. Development 121, 1139–1150 (1995).
Tash, J. S., Kakar, S. S. & Means, A. R. Flagellar motility requires the cAMP-dependent phosphorylation of a heat-stable NP-40-soluble 56 kd protein, axokinin. Cell 38, 551–559 (1984).
Wertheimer, E. et al. Compartmentalization of distinct cAMP signaling pathways in mammalian sperm. J. Biol. Chem. 288, 35307–35320 (2013).
Kamenetsky, M. et al. Molecular details of cAMP generation in mammalian cells: a tale of two systems. J. Mol. Biol. 362, 623–639 (2006).
Wickett, N. J. et al. Phylotranscriptomic analysis of the origin and early diversification of land plants. Proc. Natl. Acad. Sci. USA 111, E4859–4868 (2014).
Delwiche, C. F. & Cooper, E. D. The evolutionary origin of a terrestrial flora. Curr. Biol. 25, R899–910 (2015).
Renzaglia, K. S. & Garbary, D. J. Motile gametes of land plants: diversity, development, and evolution. Crit. Rev. Plant Sci. 20, 107–213 (2001).
Ishizaki, K., Nishihama, R., Yamato, K. T. & Kohchi, T. Molecular genetic tools and techniques for Marchantia polymorpha research. Plant Cell Physiol. 57, 262–270 (2016).
Ishizaki, K. et al. Development of gateway binary vector series with four different selection markers for the liverwort Marchantia polymorpha. PLoS One 10, e0138876 (2015).
Kubota, A., Ishizaki, K., Hosaka, M. & Kohchi, T. Efficient Agrobacterium-mediated transformation of the liverwort Marchantia polymorpha using regenerating thalli. Biosci. Biotechnol. Biochem. 77, 167–172 (2013).
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F. & Higgins, D. G. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882 (1997).
Kumar, S., Stecher, G. & Tamura, K. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874 (2016).
Le, S. Q. & Gascuel, O. An improved general amino acid replacement matrix. Mol. Biol. Evol. 25, 1307–1320 (2008).
Acknowledgements
Special thanks to Dr. Masayuki Ohmori, professor emeritus at the University of Tokyo, for his encouragement and suggestions about this study. We also thank Dr. Ryuichi Nishihama, Kyoto University, for his support in searching for CAPE orthologues in charophytes.
Author information
Authors and Affiliations
Contributions
M.K. and F.T. designed the research. N.S. and T.K. searched the databases and assisted in molecular manipulations of M. polymorpha. Y.U. and C.Y. performed the GUS staining. M.K., M.O. and Y.T. cloned the MpCAPE gene, made protein expression strains and performed protein purification and enzymatic assays. T.N., H.S., S.O., F.T. and M.K. cloned the CbCAPE and AcCAPE genes. F.T. performed phylogenetic analysis. M.K. analyzed the data and wrote the manuscript.
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Electronic supplementary material
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Kasahara, M., Suetsugu, N., Urano, Y. et al. An adenylyl cyclase with a phosphodiesterase domain in basal plants with a motile sperm system. Sci Rep 6, 39232 (2016). https://doi.org/10.1038/srep39232
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep39232
This article is cited by
-
Plant adenylate cyclases have come full circle
Nature Plants (2023)
-
An Arabidopsis Linker Histone-Like Protein Harbours a Domain with Adenylyl Cyclase Activity
Plant Molecular Biology Reporter (2023)
-
Characterization of the cAMP phosphodiesterase domain in plant adenylyl cyclase/cAMP phosphodiesterase CAPE from the liverwort Marchantia polymorpha
Journal of Plant Research (2022)
-
Distribution of adenylyl cyclase/cAMP phosphodiesterase gene, CAPE, in streptophytes reproducing via motile sperm
Scientific Reports (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.